This is an Application Brief and does not contain a detailed Experimental section.
This application brief demonstrates to enhance capabilities of the UNIFI Scientific Information System to facilitate a user’s review of raw LC-MS data from UPLC-MS or UPLC-MSE peptide mapping experiments.
User verification of results is simplified, laboratory practices can be standardized, and time between data collection and development decisions will be reduced.
Peptide mapping is a fundamental tool for the initial characterization of a biotherapeutic protein, and for monitoring that protein for new structural variants and altered levels of known product variants. The execution of such studies with high-resolution MS detection creates an information-rich pool of molecular data that can be challenging to mine for key results.
Software workflows designed for UPLC-MS peptide map analysis, such as BiopharmaLynx for MassLynx Software and the UPLC-MS peptide mapping workflow within Waters’ Biopharmaceutical System Solution with UNIFI, can dramatically reduce data processing time and prevent manual review of data sets.
However, laboratory SOPs and the desire for user verification of key findings often result in the need to go beyond the use of the processed results generated by these tools. Analysts are often required to delve into the raw chromatographic and spectral data to verify information. The UNIFI Scientific Information System has been developed to manage this biopharmaceutical workflow need, and includes enhanced functionality to support such user verification workflows.
A UPLC-MSE peptide map (120 min gradient) was acquired for the Enolase I protein from S. cerevisiae. Data were processed using the UNIFI UPLC-MS peptide mapping workflow using typical parameters (< 10 ppm mass error, assignment of tryptic, semi-tryptic, and single missed cleavage peptides, with the possibility of Asn deamidation, and Met oxidation).
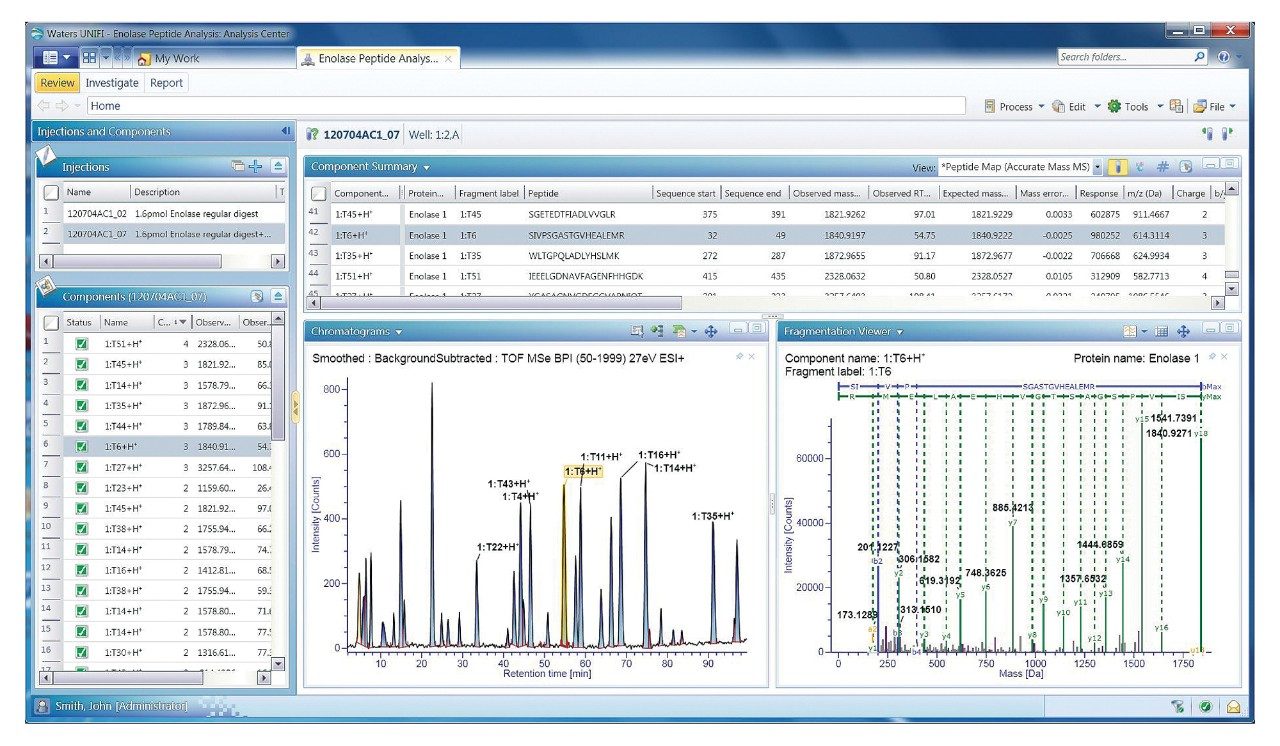
A subset of the processed results for the map is displayed as a screen-captured image of UNIFI’s Review tab for this data set (Figure 1). The Review tab provides interactive navigation of processed results between the main data table and various graphical display windows. Here, processed data for the selected T6 tryptic peptide is highlighted on the base peak intensity (BPI) chromatogram (~ 54.75 min) in the Chromatograms window, and the associated MSE spectral data is displayed in the Fragmentation Viewer window. Along with tabular results, such information can be used for rapid confirmation that automated peak assignments were correctly generated.
For unexpected results, the user can dive deeper into the raw data to verify qualitative and quantitative findings in the processed data. Opening the Investigate tab (Figure 2) reveals areas for graphical and tabular display of chromatographic and mass spectral results directly from the raw UPLC-MS data.
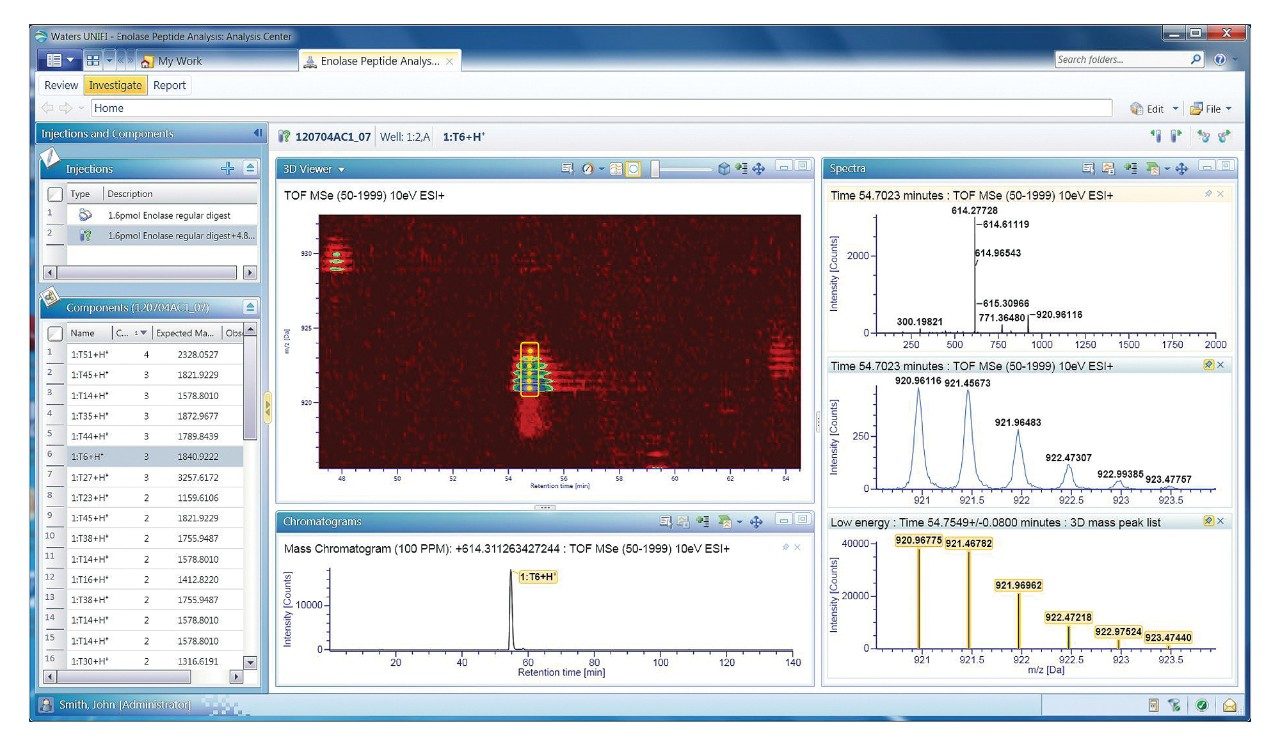
These views support open-ended investigations driven by the user, e.g., selecting masses for data extraction, summing spectra, applying background corrections, etc. Conversely, processed components are available to enable component-driven analyses of the raw data. For simplification of this figure, the same T6 peptide was selected for further investigation.
The ability of a scientist to have free reign for the review of raw data underlying a processed data set is a fundamental requirement for software applied for the analysis of complex biotherapeutic UPLC-MS data. The capabilities of the UNIFI Scientific Information System go beyond this basic requirement, and facilitate the intelligent generation of meaningful raw chromatographic and mass spectral data views, using values derived from detected map components. User verification of results is simplified, laboratory practices can be standardized, and the time between data collection and better development decisions will be reduced.
720003842, January 2011