
This is an Application Brief and does not contain a detailed Experimental section.
This application brief demonstratesdemonstrate that the ACQUITY QDa Detector is able to detect and report mass information for a diverse set of oligonucleotides.
The ACQUITY QDa Detector is a robust solution for adding cost-effective in-line mass detection to existing UV-based workflows in oligonucleotide analyses.
Chemical modifications to the phosphodiester backbone are commonly incorporated into synthetic oligonucleotides to increase stability and safety in vivo against endo- and exonucleases as well as for improving efficacy through increased cellular uptake and binding properties. While the synthesis process of oligonucleotides is well controlled, analytical characterization of synthetic products must be carried out prior to use in therapeutic applications to ensure product purity and identity.
Mass spectrometry (MS)-based techniques have increasingly been deployed in oligonucleotide analyses as an efficient means to identify process related impurities as well as providing orthogonal product confirmation. MS-based techniques that add value through improved productivity and can be adapted to existing workflows with minimal effort are highly desirable.
The ACQUITY QDa Detector provides a simple and cost-effective solution for adding mass detection into existing UV-based workflows as an in-line mass detector (Figure 1). As part of a robust solution when implementing new technologies, demonstrating method compatibility over a broad set of conditions typical of industry is ideal. To this end, Ion Pairing Reversed Phase Liquid Chromatography (IP RPLC) is one of the prevalent techniques in the analysis of synthetic oligonucleotides due in part to its enhanced performance over conventional RPLC methods and its ability to incorporate MS compatible reagents and buffers. Recently it was demonstrated that the ACQUITY QDa Detector is able to detect and report mass information for oligonucleotides over a wide molecular weight range.¹ To demonstrate that, the ACQUITY QDa Detector has applicability to a broader set of analytes beyond polyT standards, three types of oligonucleotides (siRNA, fully thioated or PS, and 2’OMe modified) representing the most commonly used in therapeutic settings were analyzed.

The Waters OST BEH C18 Column (130 Å, 1.7 µm, 2.1 mm x 50 mm) was selected for this work based on the exceptional sample resolution and superior column life it can provide for oligonucleotide separations when using high pH mobile phases. As shown in Figure 2, the n-1 and n+1 impurities, confirmed via Δ mass using ProMass by Novatia, were well resolved from the target product for all three oligonucleotide sample types using similar gradient profiles.² The total ion chromatogram (blue trace) shows a strong correlation with the optical data (red trace) for all three sample types, suggesting that the ACQUITY QDa Detector has broad applicability over diverse sample sets of oligonucleotides. Furthermore, as an in-line detector the ACQUITY QDa Detector enables orthogonal product confirmation via mass using the MaxEnt Deconvolution algorithm within the MassLynx Software (inset figure), demonstrating its ability to increase productivity and confidence of data analysis in the manufacturing process of synthetic oligonucleotides.
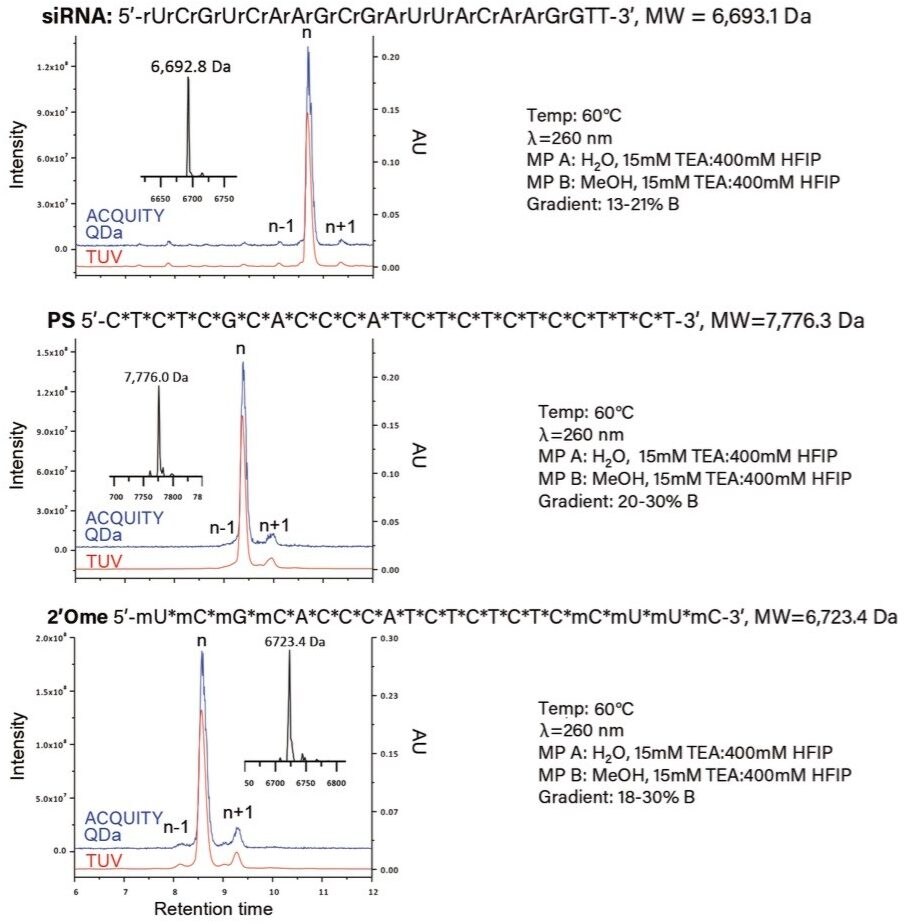
Integration of new technologies that add value and can be implemented into existing workflows with minimal effort are highly desirable. From this work it has been demonstrated that the ACQUITY QDa is able to detect and report mass information for a diverse set of oligonucleotide samples. Collectively, these results establish the ACQUITY QDa as an ideal addition to increase productivity and confidence of data analysis for routine identification and purity assessments in the manufacturing process of synthetic oligonucleotides.
720005972, April 2017