
This application note describes the use of Waters ACQUITY UPLC coupled with Xevo G2 QTof, along with POSI±IVE Software and the MassFragment tool, to screen samples from the gullet of a red kite carcass suspected of poisoning by pesticides, and to identify which pesticides were used.
We demonstrate the detection and identification of pesticide poisons ingested by a protected bird of prey.
As fragile ecosystems struggle to survive the impact of human domination of the environment, wildlife protection becomes increasingly important. While it is always preferable to safeguard living specimens in their native habitats, sadly, it is sometimes necessary to deal with the consequences of human interaction with vulnerable animals. Here we describe the use of a ToF screening approach in an incident of pesticide poisoning of a protected bird of prey.
The red kite (family: accipitridae, latin name: milvus milvus), shown in Figure 1, is a bird of prey that belongs to the same family as hawks, vultures, and eagles. This species has approximately 18,000 to 24,000 current breeding pairs in Europe, with around two thirds of this population found in Germany, and further significant populations in France and Spain. Up until the mid-19th century, red kites were persecuted extensively as vermin in the U.K.. The species was brought back from the brink of extinction by an on-going conservation effort. There are now just over 1,000 breeding pairs in the U.K., mainly located in central Wales, along the spine of central England and at various sites in Scotland.1, 2

The red kite is primarily a scavenger that feeds on worms, small mammals, and carrion. Its feeding habits make it particularly susceptible to pesticide poisoning, either accidental – when it feeds on creatures that have previously been killed by pesticides; or intentional – when people spike pesticides into carrion, either to kill animals such as foxes and crows, or to target the birds themselves.
In the U.K., the red kite is protected under the Wildlife and Countryside Act of 1981, and, under Schedule 1, Part I, of this act, they are “protected by special penalties”.3 The birds are afforded additional, wider protection in Scotland, as a result of the Nature Conservation (Scotland) Act of 2004.4 If red kite carcasses are discovered by police or wildlife protection officers, and pesticide poisoning is suspected, they are often brought to SASA (Science and Advice for Scottish Agriculture – a division of the Scottish government). Here, samples are analyzed to identify the cause of death and, if necessary, the particular type or types of pesticide used.
This application note describes the use of Waters ACQUITY UPLC coupled with Xevo G2 QTof, along with POSI±IVE Software and the MassFragment tool, to screen samples from the gullet of a red kite carcass suspected of poisoning by pesticides, and to identify which pesticides were used. We were able to demonstrate the unequivocal detection and identification of the pesticide poisons ingested by the red kite.
|
LC system: |
ACQUITY UPLC |
|
Runtime: |
5.00 min |
|
Column: |
ACQUITY BEH C18 1.7 μm, 2.1 x 50 mm |
|
Column temp: |
45oC |
|
Mobile phase A: |
10 mL of 1 M aqueous ammonium acetate solution and 990 mL water |
|
Mobile phase B: |
10 mL of 1 M aqueous ammonium acetate solution and 990 mL methanol |
|
Flow rate: |
0.6 mL/min |
|
Injection volume: |
3.0 μL |
|
MS system: |
Xevo G2 QTof |
|
Ionization mode: |
ESI positive |
|
Analyzer: |
Resolution mode |
|
Scan time: |
0.1 s |
|
Capillary voltage: |
1.0 kV |
|
Sampling cone: |
30 |
|
Source temp; |
120 °C |
|
Desolvation temp: |
550 °C |
|
Desolvation gas: |
1000 L/hr |
|
Cone gas: |
50 L/hr |
|
Mass range: |
50 to 1000 m/z |
|
MSE conditions |
|
|---|---|
|
Low energy: |
6 |
|
High energy ramp: |
25.0 – 35.0 |
|
Compound: |
Leucine enkephalin |
|
Masses: |
m/z 556.2771 and m/z 278.1141 |
|
Flow rate: |
20 μL/min |
|
Capillary voltage: |
3.0 kV |
|
Collision energy: |
21 |
The gullet contents were removed from the red kite carcass and 2.0 g were extracted into 5 mL of ethyl acetate.
A 1 mL aliquot was then solvent exchanged into methanol and made up to 400 mL, i.e. 0.4 g of gullet content extract in 400 mL.
This sample was passed through a 0.2 μm syringe filter with no further cleanup prior to analysis.
Mobile phase gradient is detailed in Table 1.
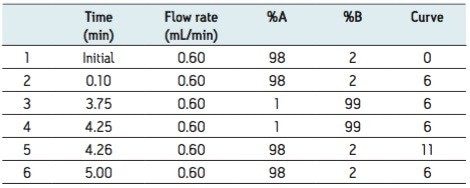
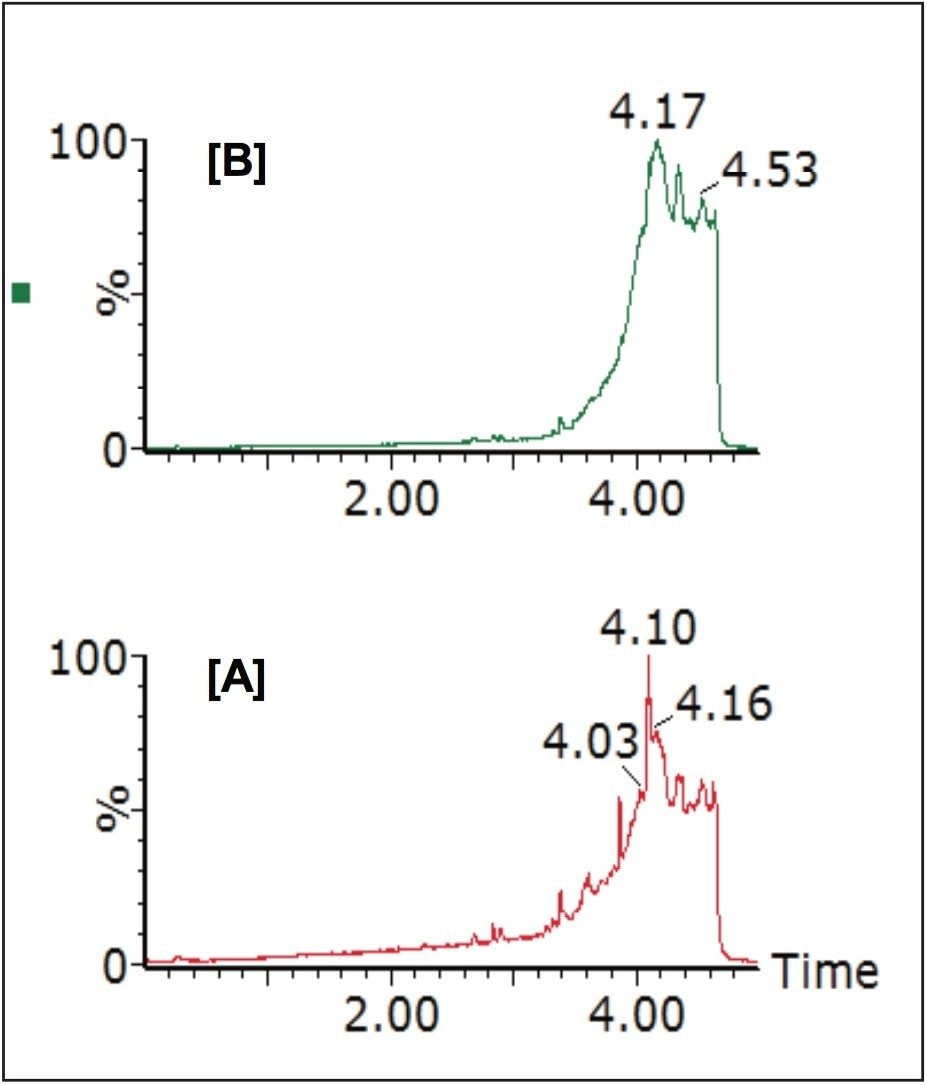
The generic screening method given above was used to screen extracted gullet contents of the red kite for pesticide residues. The low energy MSE precursor ion total ion chromatogram (TIC) and the high energy MSE fragment ion TIC acquired from this screening analysis are shown in Figure 2.
Matrix-matched standards were not available for the red kite sample; however, the sample data were processed using POSI±IVE Software with pesticide solvent standards in order to provide identification and help assess the magnitude of the compounds that poisoned the red kite.
Figure 3 shows the ChromaLynx XS Identify results browser from POSI±IVE data processing. Here, 585 compounds were targeted from which two were automatically found and identified as carbofuran and carbosulfan. Data for both the MSE low energy precursor ions and the high energy fragment ions are displayed. The accuracy of the exact mass ions, as shown for carbosulfan (precursor ion: m/z 381.2212, fragment ion: m/z 118.0690) with ΔM values of +0.2 mDa and -0.3 mDa respectively, provides added confidence that the results are correct.
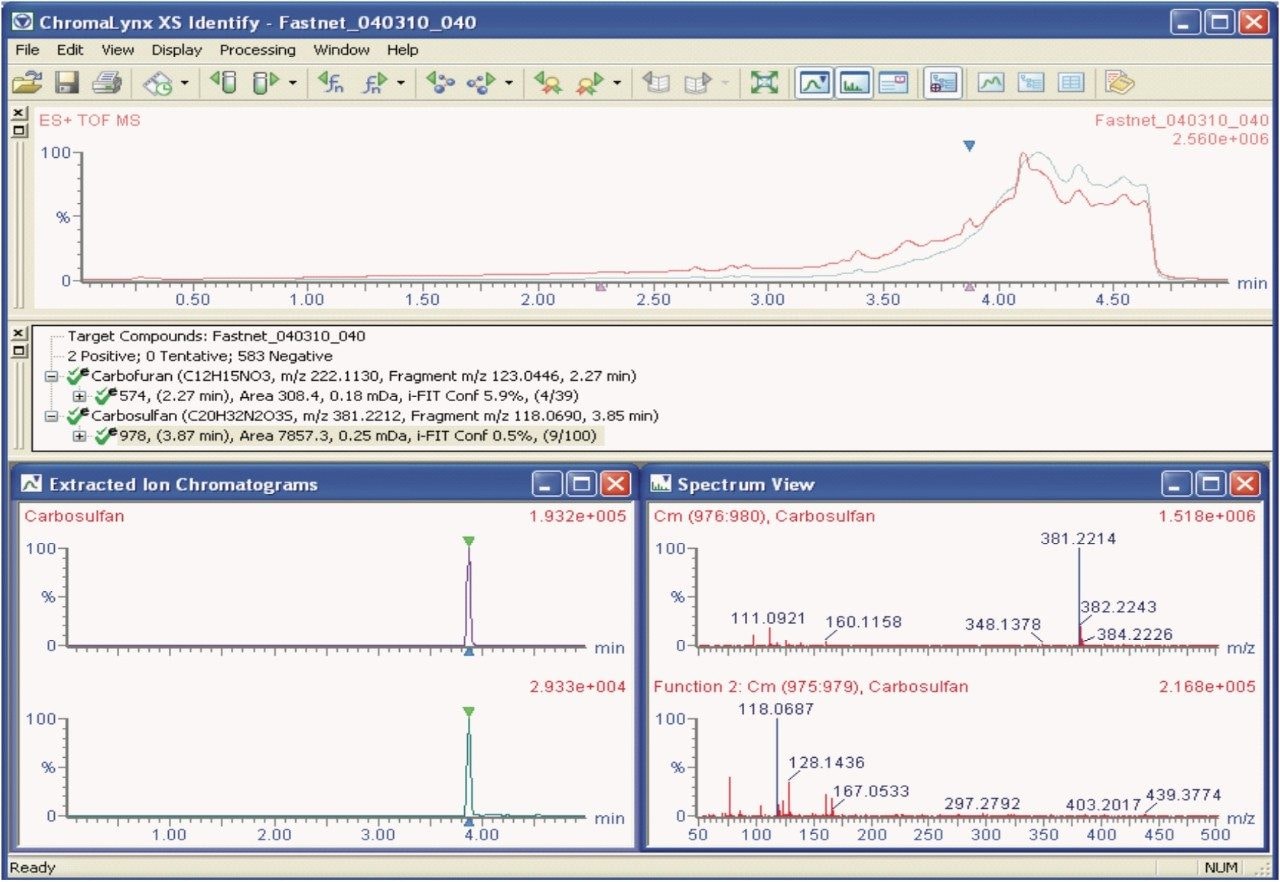
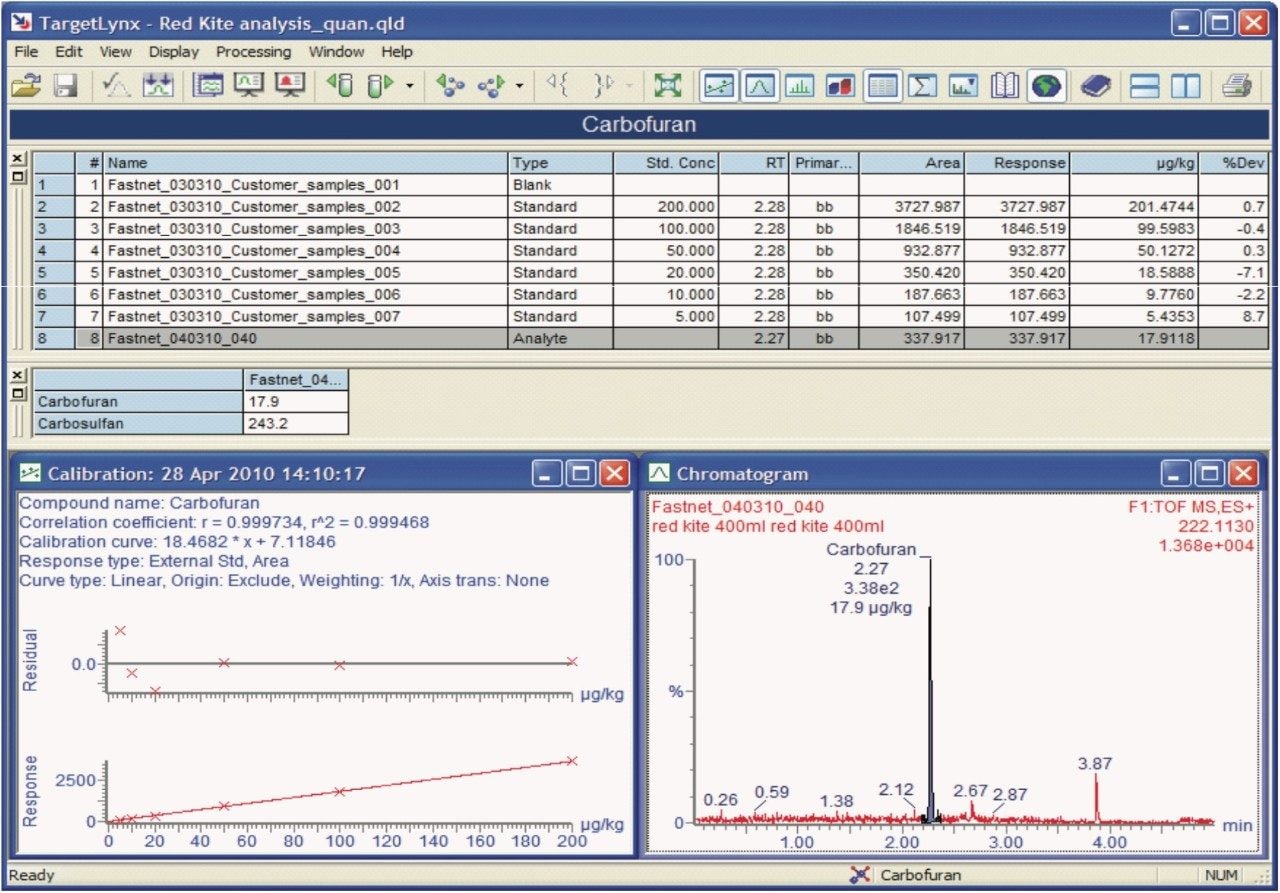
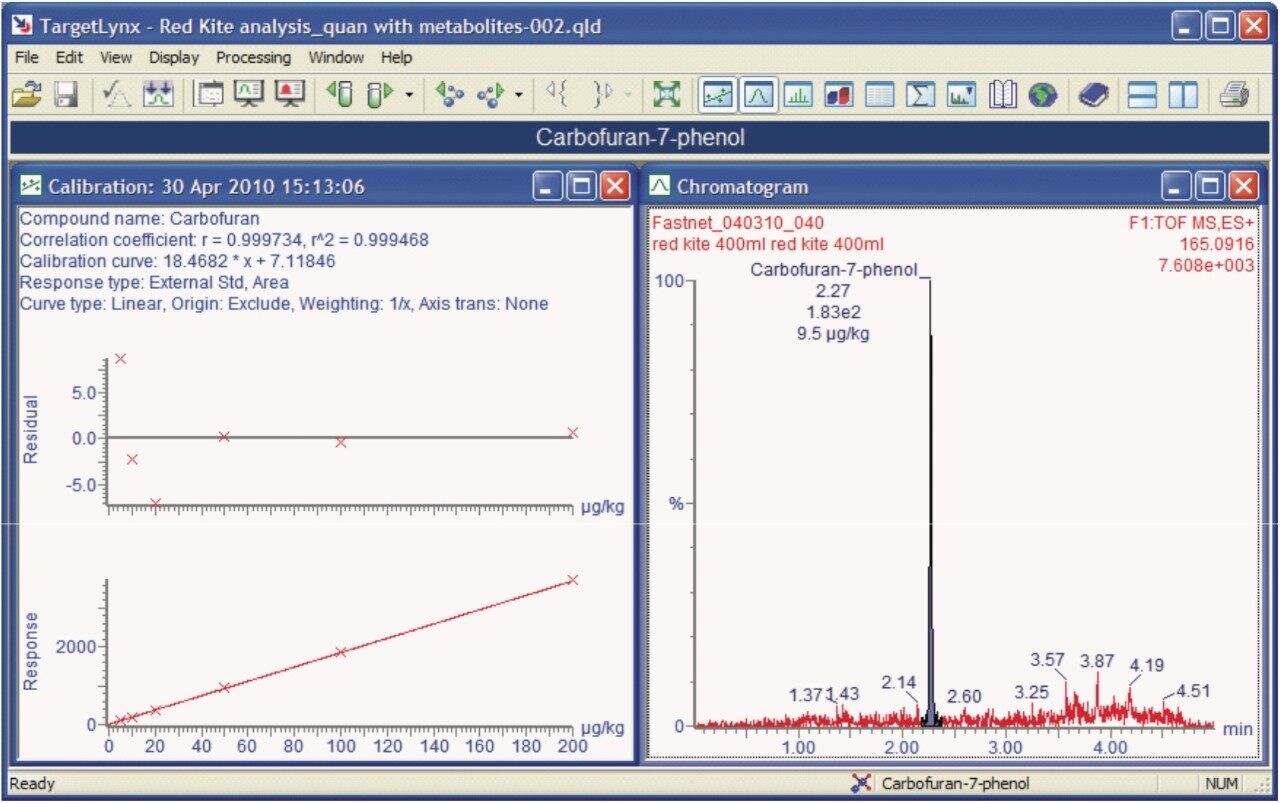
Figure 4 shows the TargetLynx Quantify Browser from POSI±IVE data processing. Here pesticide solvent standards have been used to quantify the identified pesticide poisons.
Once the toxic compounds had potentially been identified as carbofuran and carbosulfan, additional metabolites of these two pesticides, previously identified in work by Soler, et al.5, were included in the targeted screening database, and the data were re-processed using POSI±IVE Software. Figure 5 shows a section of the targeted compound database used, with the added metabolites and parent compounds highlighted.
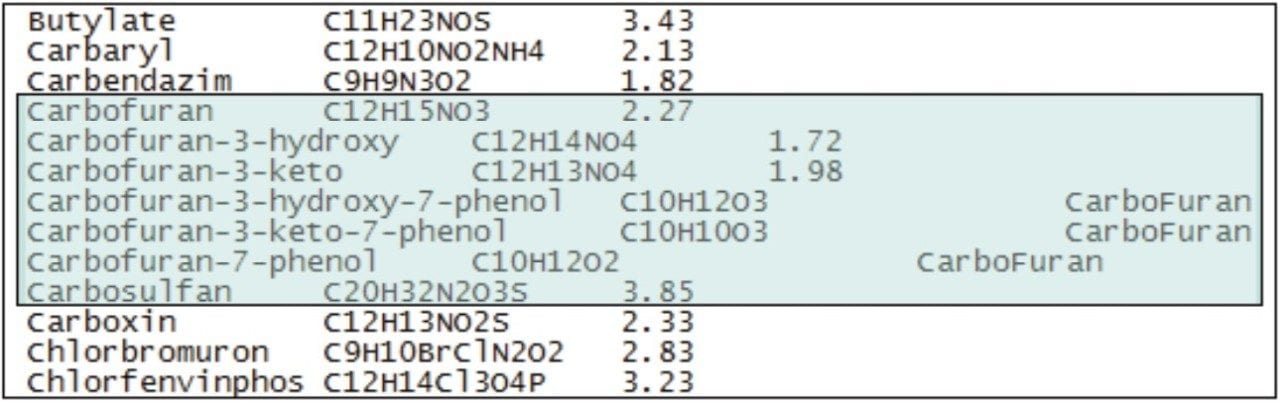
Standards were not available for some of the metabolites of interest; however, POSI±IVE provides the opportunity to identify a similar compound on the list for use as a standard from which to quantify. In Figure 5, 3-hydroxy-7-phenolcarbofuran, 3-keto-7-phenolcarbofuran, and 7-phenolcarbofuran, if present in the sample, would each be quantified using the calibration curve for carbofuran.
After re-processing, the metabolite 7-phenolcarbofuran was also identified and quantified using the solvent calibration curve for carbofuran, as shown in Figure 6.
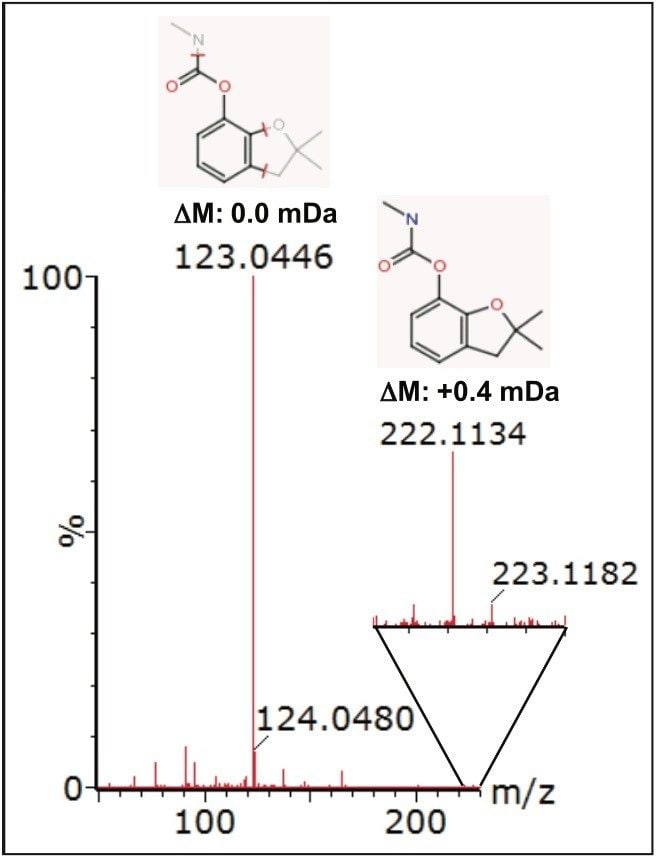
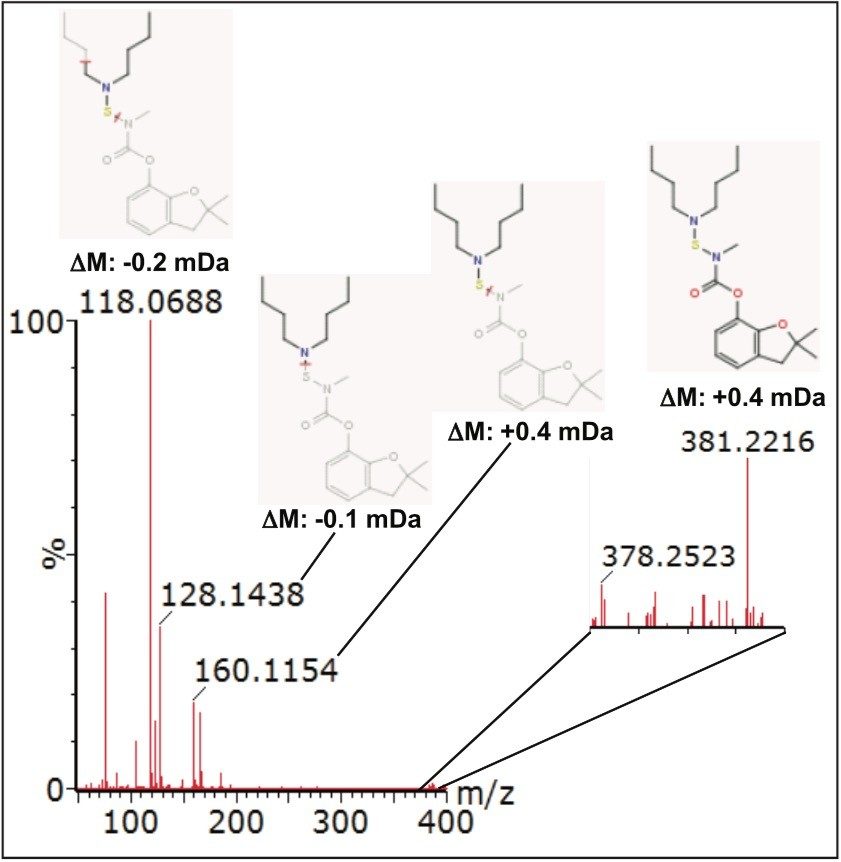
Further compound confirmation was carried out using the MassFragment tool. Structures were assigned to the MSE fragment ion spectra acquired from the relevant extracted ion chromatograms (XIC), based on accurate and precise exact mass data. Figure 7 shows MassFragment-assigned structures for the fragment ions seen at 2.27 min, and Figure 8 shows similar information for the fragment ions acquired at 3.87 min.
With thanks to SASA for providing extracted samples and pesticide standards.
720003470, May 2010