Effects on Endogenous Metabolites Following the Administration of Methapyrilene: A Data-Dependent Acquisition Workflow Using the Xevo™ MRT Mass Spectrometer
Lee A. Gethings,a Susan Abatiello,b Jennifer Seymour,c Terri Sosienski,c Jeff Finch,c Jeremy Felton,c Donald Laudicina,c Richard Lock,a Robert S. Plumb,c Ian D. Wilsond
a Waters Corporation, Wilmslow, United Kingdom
b Immerse Lab, Cambridge, MA, United States
c Waters Corporation, Milford, MA, United States
d Imperial College, London, United Kingdom
Published on August 28, 2025
Abstract
Endogenous metabolomic changes observed with the administration of drugs (e.g. gefitinib) have previously been reported.1 These effects, termed “pharmacometabodynamic” effects, are the dynamic, time-related, and reversible changes in metabolic phenotypes resulting from the pharmacological effects of a drug (or other bioactive substance) on the metabolome. Methapyrilene (MP), N,N-dimethyl-N'-pyridin-2-yl-N'-(thiophen-2-ylmethyl)ethane-1,2-diamine, is a 2-thiophene-based H1-receptor antagonist and has been attributed to drug induced liver injury (DILI).2 Using a rat model, the potential pharmacometabodynamic behavior is highlighted for various doses of MP over a time course of 6 days. Implementing the Xevo MRT Mass Spectrometer provides excellent mass accuracy, and high mass resolution and sensitivity for metabolomic studies.
Benefits
- Demonstration of the Xevo MRT Mass Spectrometer (MS) workflow for the analysis of urine extracts from a drug dosed study involving methapyrilene.
- Data dependent acquisition (DDA) has been used for data collection, providing a high level of fragmentation information to confidently identify compounds of interest.
- Combining a DDA approach with high throughput chromatography is shown to consistently yield high quality, highly confident data.
- Integration of data with third-party software, MS-DIAL, provides a flexible workflow for data processing and compound identification.
Introduction
DDA has long been established for providing highly specific MS/MS information, which assists with structural elucidation and hence metabolite identification. The speed of the Xevo MRT Mass Spectrometer allows DDA data to be acquired at 50 and 100 Hz, MS and MS/MS respectively and therefore being particularly amenable to fast chromatographic methods. The study outlined emphasizes high quality DDA data combined with fast chromatography, enabling highly confident metabolite identifications to be gained.
Experimental
Study Overview
Wistar rats (n=18) were dosed with MP formulated at either 0, 5 or 15 mg/mL (vehicles) for a 150 mg/kg dose group. MP was administered orally (PO) once/day by gavage for 5 days at 10 mL/kg. Urine was collected over a 16 hour period on predose (day 0-1), day 2, day 3, and from day 5 to day 6. The MP dosing studies were all conducted at Evotec SAS (Toulouse, France). Ethics were based according to National and EU guidelines.
Sample Preparation for LC-MS
Urine samples were firstly protein precipitated using 10 µL urine with 40 µL ACN:MeOH (90:10 v/v). Samples were then vortexed prior to storing at -20 °C for 2 hours. Following incubation, samples were further vortexed and centrifuged at 25,000 g for 5 minutes. The resulting supernatant was then diluted 1:5 (v/v) with distilled water and transferred to LC-MS total recovery vials (p/n: 186002805).
LC-MS Data Acquisition
LC-MS data were collected using an ACQUITY™ Premier UPLC™ System coupled to a Xevo MRT Mass Spectrometer. The reversed-phase (RP) chromatographic separation consisted of a 5.0 minutes method (injection to injection) using a CORTECS™ C18, 1.6 µm, 2.1 x 50 mm analytical Column (p/n: 186007114). The LC solvents comprised of mobile phase A (0.1% formic acid containing 1 mMol ammonium formate) and mobile phase B (95:5 ACN:Water (v/v), 0.1% formic acid, 1 mMol ammonium formate) being ramped from 50% to 99% over 5 minutes using a flow rate of 0.6 mL/min. The gradient started from 0.1% to 15% (mobile phase B) over 1.5 minutes, 50% at 3 minutes and 99% at 4 minutes. The solvent composition was held at 99% (mobile phase B) for 0.5 minute and re-equilibration at initial conditions by 5 minutes, ready for the subsequent injection.
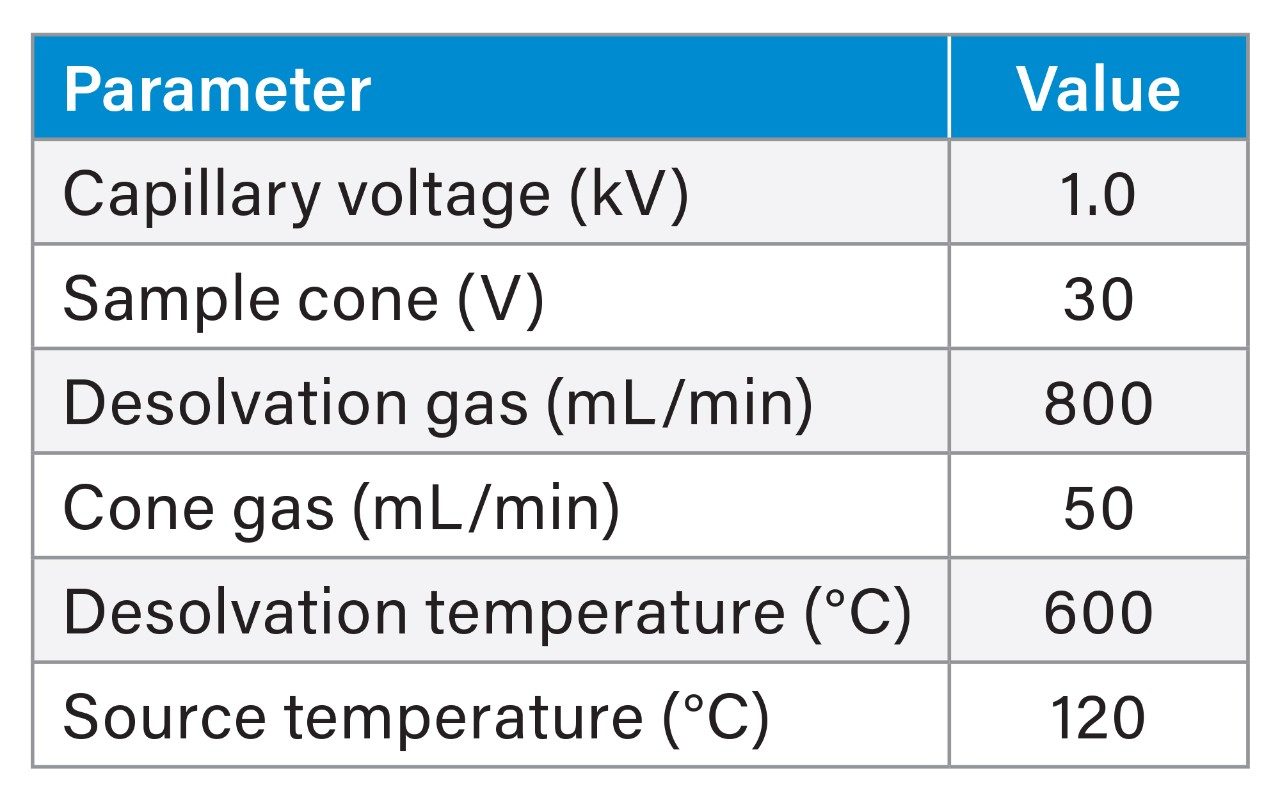
The Xevo MRT Mass Spectrometer was configured with the source settings as outlined in Table 1. MS data were collected using DDA mode of operation, which consisted of scan rates of 20 Hz (MS) and 50 Hz (MS/MS). The number of components selected for MS/MS were 8 with a dynamic exclude applied of 10 mDa with a 10 second retention time tolerance.
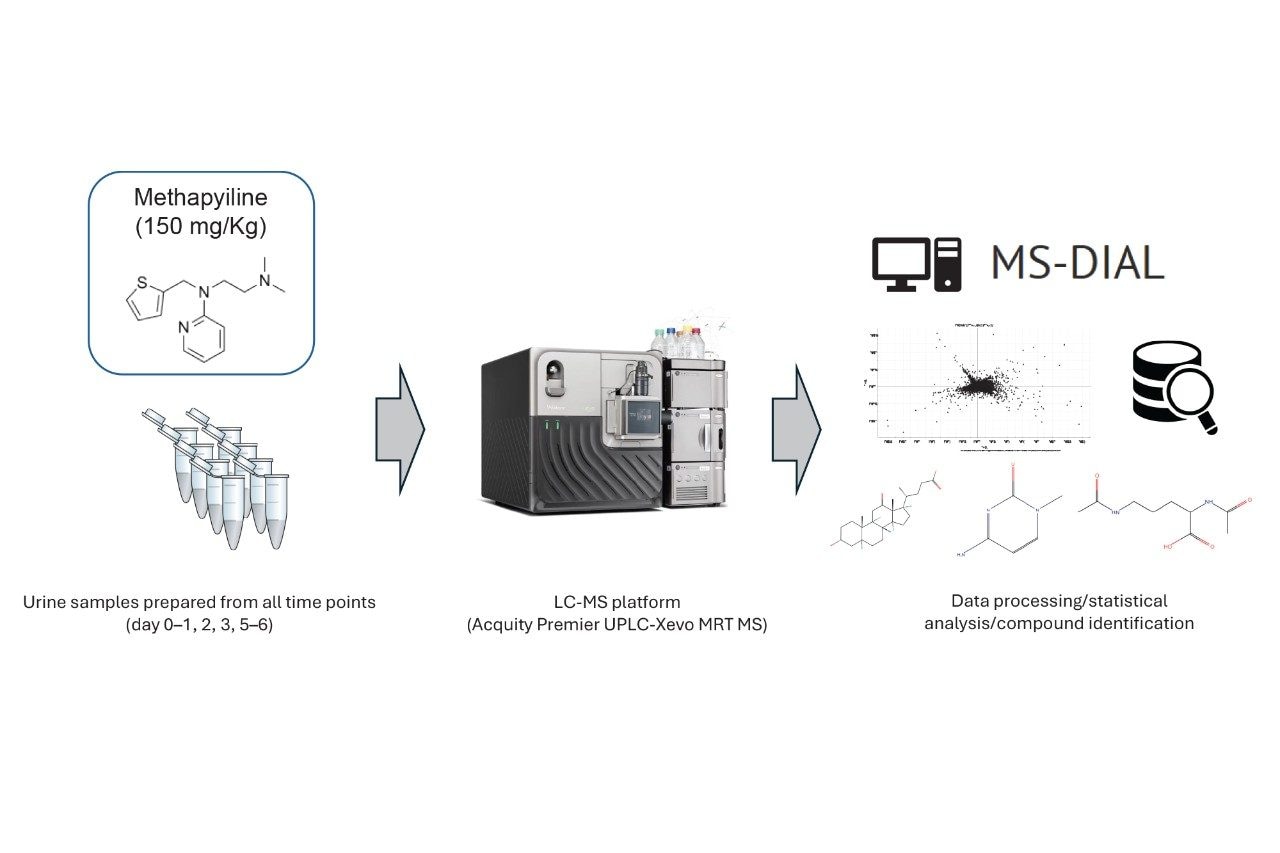
Data Processing
The data were converted to mzML format during the data acquisition process. MS-DIAL3 was used to peak pick, normalize, and align the data. Statistical analysis and compound identification were also performed within MS-DIAL. Compound identifications were obtained by searching against a spectral library comprised of authentic standards.
Results and Discussion
Data were acquired for all the rat urine samples using the Xevo MRT Mass Spectrometer, operating in DDA mode. The subsequent data were peak picked, aligned, and normalized using MS-DIAL (Figure 1).
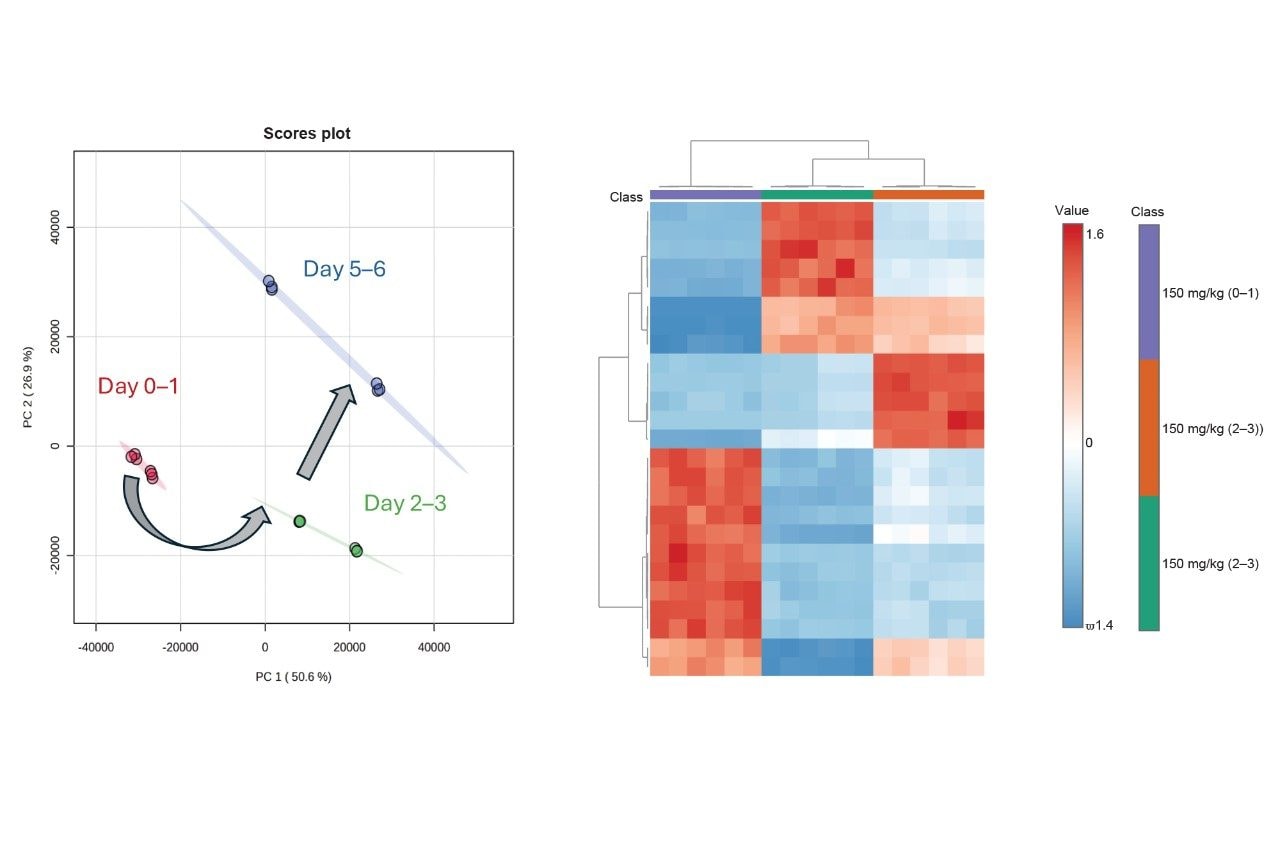
The data matrix resulting from the processing stage provided a total of 18,653 peak picked features. These were subsequently subjected to multivariate statistical analysis (MVA) to determine relevant, differential features over the study time course for compound identification (Figure 2). Prior to MVA, any features corresponding with the drug or its associated metabolites were removed from the data matrix, ensuring that observed differences were due to endogenous features. Principal component analysis (PCA) showed a change over the time course and provided evidence of cage effects between the animals. Applying Analysis of Variance (ANOVA) and plotting the data as a heatmap with hierarchical clustering, provided the top 25 features responsible for the separation observed over the three time points. These features were then searched against a compound library comprised of authentic standards (Figure 3).
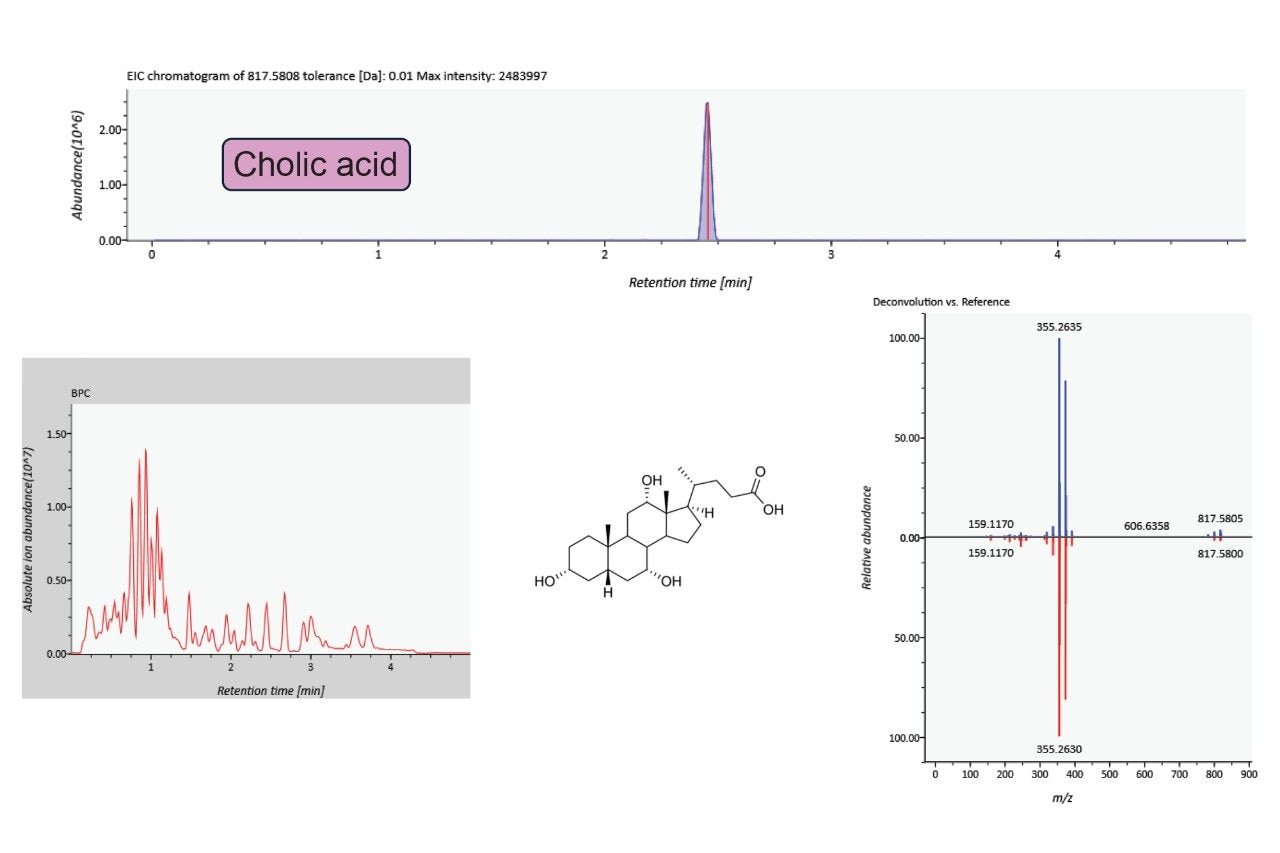
Identifications included a range of metabolites including amino acids, organic acids and nucleosides, that were confidently identified. Figure 2 serves as one example, with cholic acid being comprehensively identified as shown by the mirror plot, indicating high MS/MS coverage and excellent mass accuracy. The high mass accuracy provided by the Xevo MRT Mass Spectrometer significantly reduces the number of false positives following a database search. Comparing compound identifications based on mass tolerances of either 5 ppm (MS) and 2 ppm (MS/MS) versus 1 ppm for MS and MS/MS, the number of features mapped to potential identifications were 8869 and 4693 respectively.
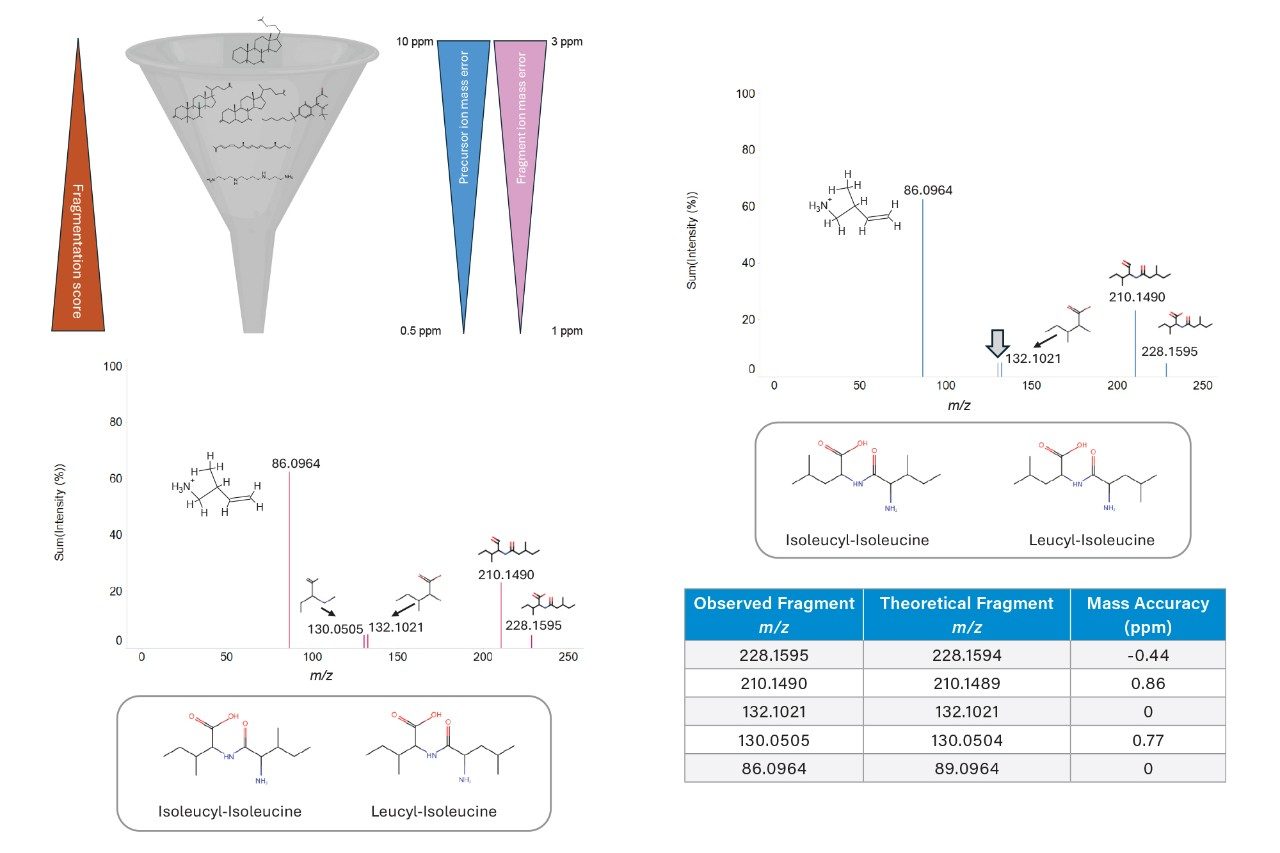
Deciphering between potential identifications relating to an individual feature is particularly challenging when conducting database searches on complex biological samples such as urine and plasma. The high coverage of MS/MS peaks combined with high mass accuracy is further highlighted in Figure 4. Database searches (HMDB based) conducted with precursor/fragment tolerances of 10 ppm/3 ppm were compared with searches using mass tolerances of 0.5 ppm/1 ppm. Based on the stringent tolerances of 0.5 ppm/1 ppm, the number of potential identifications relating to the mass feature (m/z 245.1859) reduced from 48 to 4. All four potential identifications are related to a dipeptide with a precursor mass accuracy of 0.17 ppm, and all corresponding fragments being at sub 1 ppm accuracy. Various forms of the dipeptide exist, including enantiomers (isoleucyl-isoleucine/leucyl-isoleucine and isoleucyl-leucine/leucyl-leucine) which further complicate complete identification. Inspection of the MS/MS spectra, however, shows the matching of a single peak at m/z 130.0505 allows for isoleucyl-isoleucine/leucyl-isoleucine to be the most confident compound relating to feature m/z 245.1859.
Conclusion
Confident compound identification in metabolomic and lipidomic studies is a challenging task due to highly complex samples, multiple compound conformations and access to authentic standards for ultimate identification. Mass spectrometry is an established technology for metabolite identification and as technology advances, so does the ability to confidently identify compounds of interest. Xevo MRT Mass Spectrometer operating in DDA provides highly informative MS/MS spectra with the added benefit of high mass accuracy. The experiments outlined here have shown how these attributes can be applied to a metabolomic workflow helping researchers to reduce the number of false positives reported.
References
- Molloy et al. The Pharmacometabodynamics of Gefitinib after Intravenous Administration to Mice: A Preliminary UPLC-IM-MS Study, Metabolites. 2021, 11(6):379.
- Graichen et al. Effects of methapyrilene on rat hepatic xenobiotic enzymes and liver morphology. Fundam. Appl. Toxicol. 1985 5, 165–174.
- Tsugawa et al. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nature Methods, 2015 12, 523–526.
720009003, August 2025