This is an Application Brief and does not contain a detailed Experimental section.
The goal of this application brief is to characterize complex protein digest samples using a new data dependent acquisition algorithm.
Obtain consistent results between injections and more confident results for DDA acquisitions
LC-MS/MS using data dependent acquisition has been widely employed to qualitatively characterize tryptic digests and subsequently identify the constituent proteins. While many studies have been conducted and a large number of instruments have been employed using this approach, it is well recognized to have a number of serious limitations, including under-sampling, irreproducibility, lack of in-sample dynamic range, and an inability to deal with chimericy. This has resulted in the development of alternative data independent strategies, such as LC-MSE, to address these issues. However, data dependent acquisition is important for the analysis of samples labeled with isobaric quantitation reagents (i.e. iTRAQ and TMT), which cannot be currently addressed by data independent methods. In this technology brief, we present an improved DDA method for the characterization of such proteomic samples.
This new FastDDA algorithm includes the following features:
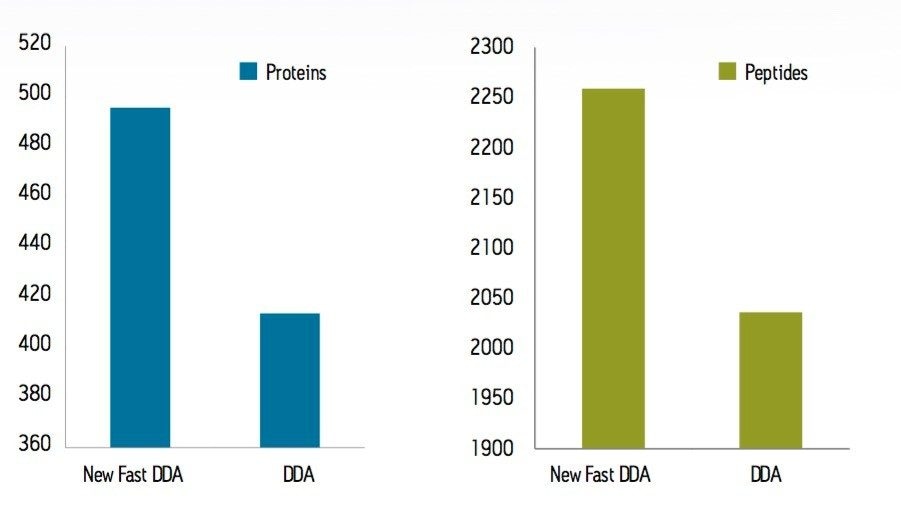
To test the performance of the new algorithm, a complex tryptic digest from the cytosolic fraction of E.coli was separated on a nanoACQUITY UPLC System with an ACQUITY UPLC BEH 1.7 µM, 75 µM x 100 mm Column. A gradient from 1% to 40% acetonitrile + 0.1% formic acid over 90 minutes was used at a flow rate of 300 nL/min. The UPLC eluent was passed directly into the NanoFlow ion source of a Xevo G2 QTof Mass Spectrometer. For all experiments, 400 ng of total protein digest were injected on column, and the FastDDA parameters that were used in these experiments are shown in Table 1. The results in terms of peptides and proteins identified for the FastDDA algorithm are shown in Figure 1. The results obtained from the FastDDA code were then directly compared to the previous MassLynx Software DDA acquisition code, as shown in Figure 2.
It can clearly be seen that both the number of peptides and proteins identified by the new FastDDA algorithm is superior.
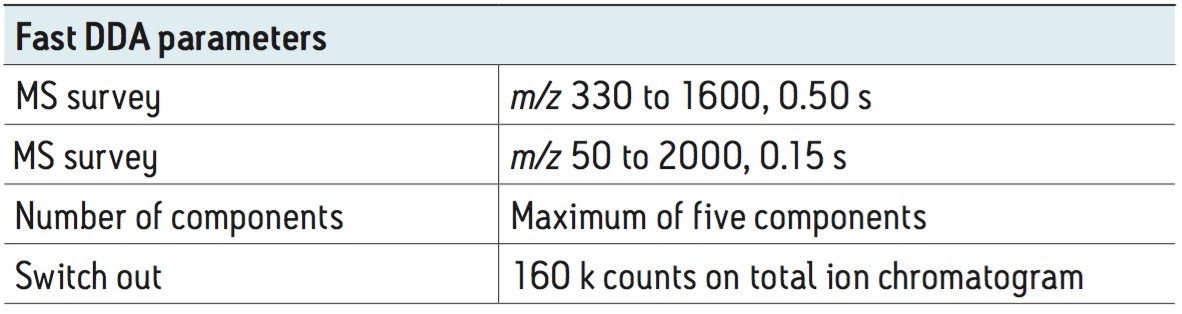
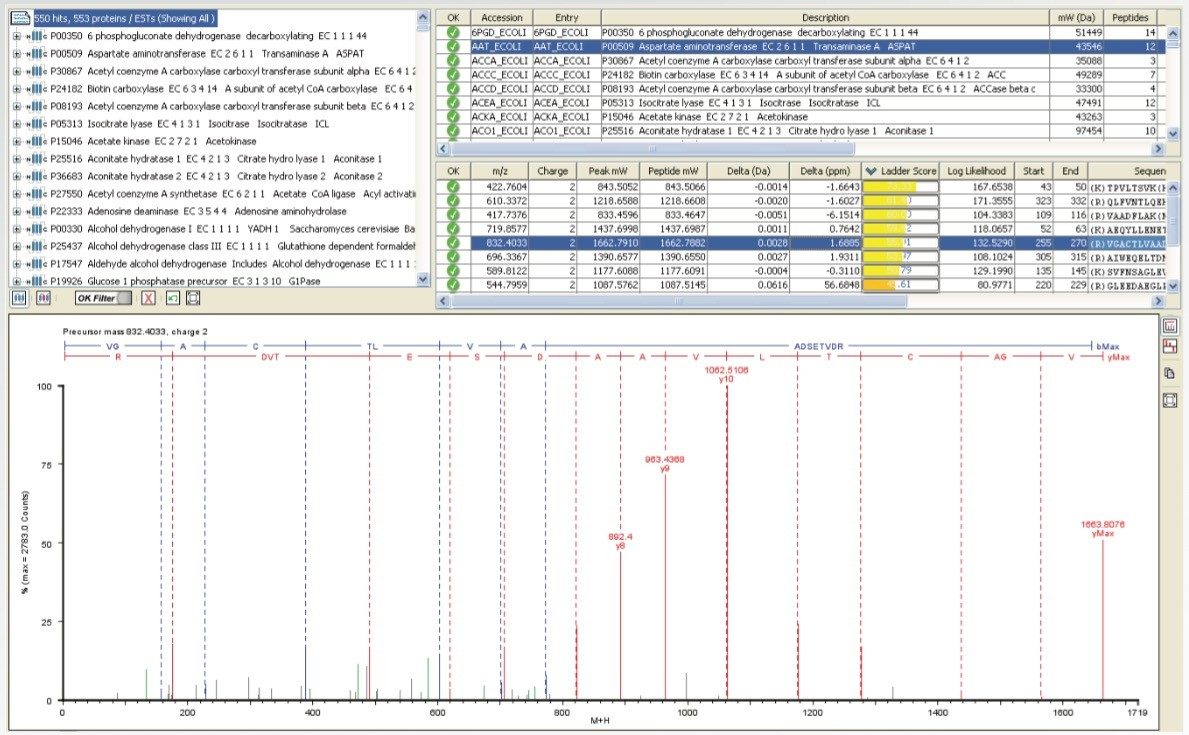
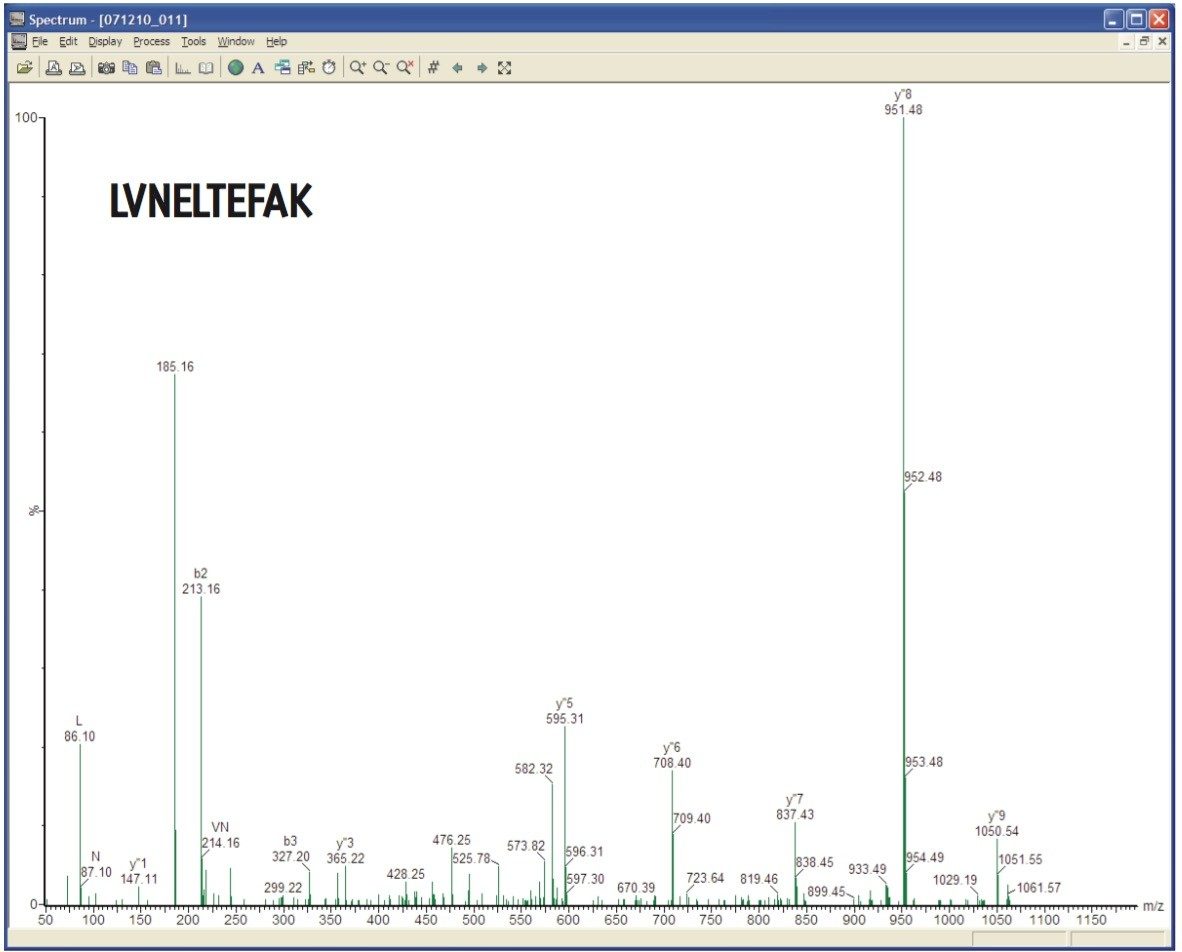
MS/MS spectra can be acquired at rates of up to 30 spectra per second, depending on the abundance of the ion. An example of this is demonstrated in Figure 3, which shows a 36 msec integration from a BSA tryptic digest. Despite the short integration time, the resolution and mass accuracy in both MS and MS/MS modes were maintained, providing highly-specific information for identification purposes.
Here we have described new FastDDA acquisition code. The benefits of the FastDDA acquisition code are that it delivers more consistent results between injections and it provides greater coverage of the peptides and proteins present. Together, these enable more confident results to be obtained for DDA acquisitions in proteomics laboratories.
720003961, May 2011