This is an Application Brief and does not contain a detailed Experimental section.
As of 2013, there were 338 new monoclonal antibody drugs in development,1 representing the largest class of biologics in drug development pipelines. Recently, LC-MS has been gaining ground as the technique of choice over ligand binding assays (LBAs) for the support of biologic drug development programs. An important component of these LC-MS workflows is the post digest purification of tryptic peptides. While SPE is commonly used in small molecule sample preparation, the same methods and guidelines do not apply to peptide extraction. Knowledge of peptide handling concerns as well as the ability to efficiently manipulate wash and elution conditions for tryptic peptides have become critical capabilities in bioanalytical labs. Historically, these labs have been dominated by work on small molecule therapies with far simpler and more straightforward sample prep and analysis methods. The expertise in peptide and protein bioanalysis, if it exists, tends to be a rare and valuable skill, held by only a few individuals.
The aim of this technology brief is to demonstrate the performance of an optimized method for tryptic peptide clean-up in protein bioanalysis.
The simple 96-well μElution format enabled clean-up and concentration of digest samples in <20 minutes, without the need for evaporation and reconstitution, preserving low levels of precious tryptic peptides
During a typical protein bioanalytical workflow, it is common to either directly digest an unpurified plasma/serum sample or to perform affinity purification followed by digestion. An affinity step may be included to isolate a class of proteins (for example, the use of Protein A/G which isolates an IgG fraction) or a specific target from the plasma/ serum sample. Depending on the degree of protein-level clean-up, which ranges from none (direct plasma digestion) to significant (specific affinity capture of a target) the concentration of resultant peptides can be extremely high. In small molecule SPE the majority of proteins (undigested, of course) pass through the extraction device on the load step. In contrast, in a protein bioanalysis workflow, 10’s of thousands of digested peptides from as high as 50–60 mg/mL starting protein concentration, are all now capable of binding to the SPE sorbent bed. This creates a significant capacity challenge. A possible solution that might occur to a “small molecule” scientist would be to use a larger sorbent bed size, for example 10, 30 or even 60 mg sorbent beds. However, these larger bed sizes require larger volumes to elute, which must then be dried down and reconstituted. The same practices, when applied in large molecule quantification, could result in significant peptide/protein losses due to adsorption during evaporation. Therefore, it is critical to avoid the dry-down step commonly associated with small molecule SPE methods.
In addition to the requirement for clear capacity guidelines for loading protein digests, a method tailored to targeted elution of tryptic peptides is also needed. A broadly applicable protocol which effectively removes excess digest reagents, digestion buffer, phospholipids, and other plasma/serum components could not only increase sensitivity by reducing matrix effects, but would also improve system robustness.
A total of 17 signature peptides (a combination of both generic and unique) were used in the development and verification of an SPE protocol, as well as an optimal device format, which specifically targets tryptic peptide clean-up during protein bioanalysis experiments. The result of this research is the ProteinWorks μElution SPE Clean-up Kit (p/n 186008304). Key attributes of the kit include: the ability to concentrate digests without evaporation, orthogonal retention mechanism for maximum specificity, and a protocol designed expressly for efficient purification of tryptic peptides. Oasis MCX was chosen as the ideal sorbent due to its strong ion exchange binding to the positively charged termini produced through cleavage at the basic residues arginine and lysine during trypsin digestion. In addition, very polar peptides are more efficiently trapped by ion exchange on the Oasis MCX plate than they would be on a traditional reversed-phase only sorbent. The μElution 96-well device format is characterized by elution in as little as 25 μL, which enables significant concentration of extracts without evaporation. The entire 96-well plate can be processed in under 20 minutes using either vacuum or positive pressure, and is amenable to automation.
Performance of the SPE Kit was evaluated using a broad range of chemically diverse representative peptides. Key peptide characteristics are summarized in Table 1.
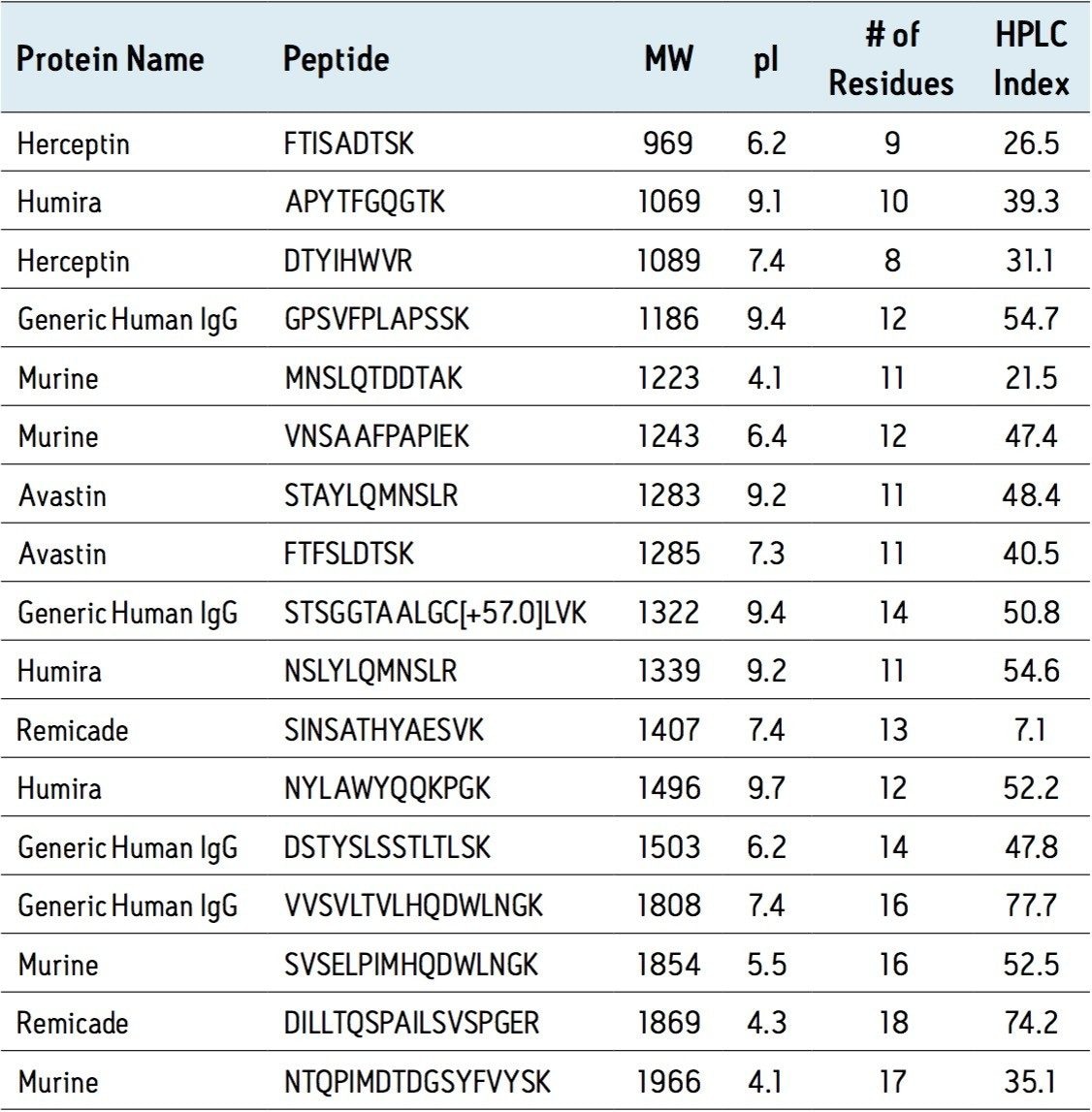
Through extensive testing and optimization, recovery was maximized for all peptides and sample loading guidelines were created. The comprehensive guidelines, which are related to starting protein content, address both direct plasma/serum digest samples and extracts resulting from affinity purification of the target protein or class of proteins. Recovery of the various peptides using the optimized protocol in the SPE Kit is summarized in Figure 1. The average peptide recovery for the antibody drugs is 104%. Peptides from the generic murine mAb IS average 84% recovery.
It is recommended that the total protein load in the digested sample on the Oasis MCX μElution plate not exceed 1.25 mg. In addition, for best performance, the minimum SPE loading volume range is 15–25 μL. The sample loading guidelines, summarized in Table 2, were designed to produce the highest sensitivity by identifying the maximum digest volume which should be loaded.

Table 2. Recommended maximum digest loading volumes for the ProteinWorks μElution SPE Clean-up Kit
* Starting volume of plasma added for protein digestion or affinity purification using the ProteinWorks eXpress Digest Kit and Protocols, with a final digestion volume of 200 μL.
**Based on a total protein content of 75 mg/mL in whole plasma/serum.
† Based on a total protein content 15 mg/mL post-generic affinity purified plasma (Protein A/G) and assuming all Affinity captured sample is used for digestion.
Plasma was spiked with infliximab, trastuzumab, adalimumab, bevacizumab, and a common murine mAb IS and then digested. The resultant tryptic peptides were isolated from digest reagents (which are washed off during loading and wash steps), phospholipids (which remain stuck to the SPE plate after peptide elution), and concentrated using the ProteinWorks SPE Clean-up Kit. Chromatographic purity of the signature peptides extracted from the plasma digest sample is demonstrated in Figure 2.
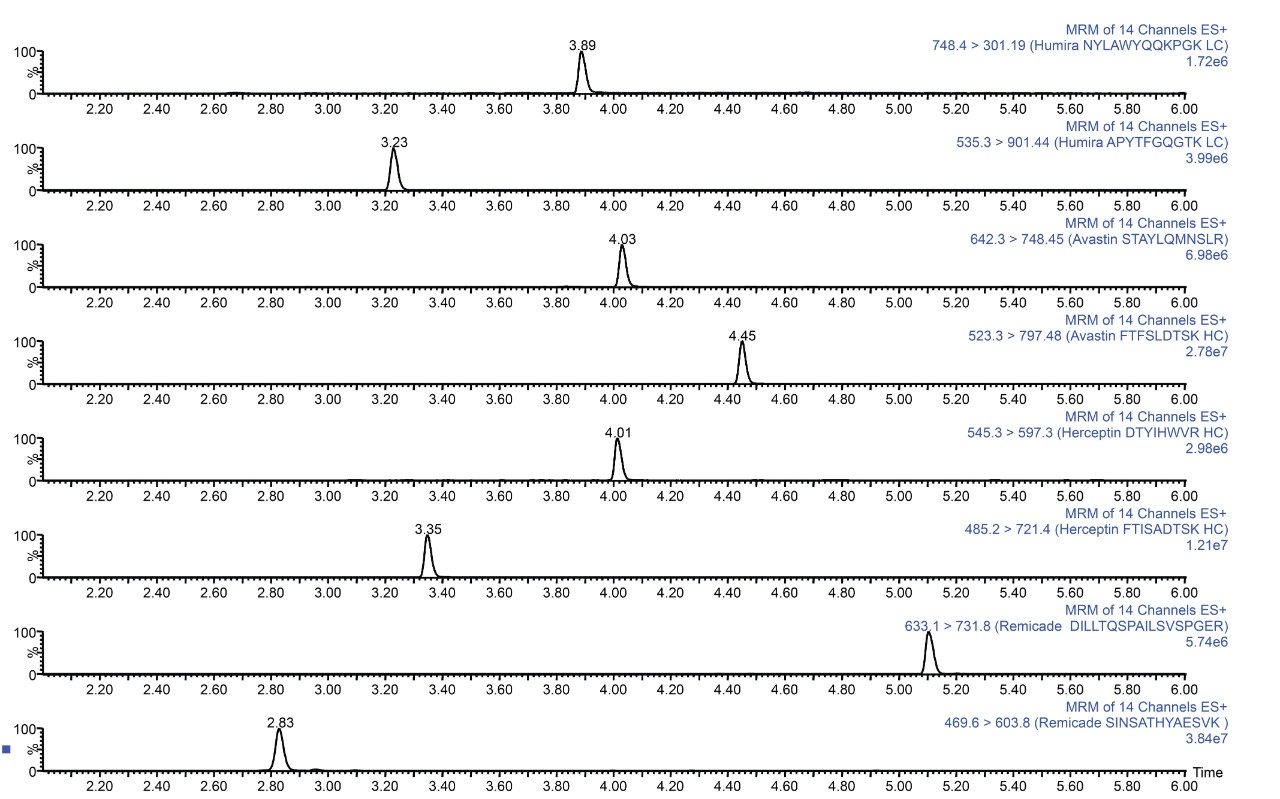
The ProteinWorks μElution SPE Clean-up Kit was successfully used to isolate peptides resulting from plasma digests of bevacizumab, trastuzumab, adalimumab, and infliximab as well as a common murine mAb internal standard. The simple 96-well μElution format enabled clean-up and concentration of digest samples in <20 minutes, without the need for evaporation and reconstitution, preserving low levels of precious tryptic peptides. Furthermore, the kit takes advantage of the orthogonal nature of mixed-mode SPE (binding tryptic peptides by ion-exchange) to provide the degree of specificity required for low-level protein quantification studies. Finally, the specific sample loading guidelines and an optimized kit-based approach allow novice users to quickly, easily, and effectively clean-up protein digests. This strategy generates data with the best possible reproducibility and highest peptide recovery for challenging protein bioanalysis studies.
720005544, November 2015