The Application of a New DDA Acquisition Method for the Enhanced Analysis of Peptides Labeled With Isobaric Mass Tags
Este es un resumen de la aplicación y no contiene una sección experimental detallada.
Abstract
To quantify proteins from complex mixtures using isobaric labelling combined with data dependent acquisition (DDA).
Benefits
- Obtain consistent quantitative measurements from isobaric labelling studies by FastDDA
Introduction
LC-MS/MS using data dependent acquisition has been widely employed to qualitatively characterize tryptic digests to identify the constituent proteins. Recently isobaric peptide tagging reagents, such as iTRAQ and TMT, have been developed that allow quantitative studies to be performed. These reagents suffer from a number of limitations such as: quantitative dampening of expression changes, limited dynamic range, and the ability to identify but not quantify peptides. Despite this, they have been utilized in the proteomics laboratory, allowing multiplexed samples to be run in a single LC/MS/MS, and are useful in small time course and sub-cellular fraction experiments. In this technology brief, we present a unique DDA method, specially designed for isobaric labeling reagents, providing optimized fragmentation patterns for the peptides and reporter ions, allowing both quantification and identification.
Results and Discussion
The new FastDDA algorithm developed for the Xevo® G2 QTof has a specific option designed for peptides labeled with an isobaric tag. The ideal fragmentation pattern for a peptide labeled with an isobaric tag contains both of the following:
1. “Y’’ and b ions related to the amino acid sequence, present at high m/z – for identification purposes
2. High-intensity of reporter ions at low m/z for quantitative purposes
These two requirements require good transmission across the m/z scale and performing these on ion-trap instruments has proven very challenging. In addition, these requirements are at odds with single collision energy during an MS/MS acquisition as Point 1 generally requires low collision energy and Point 2 requires high collision energy.
The new Fast DDA algorithm overcomes this by spending 50% of the integration time for an MS/MS acquisition at an elevated collision energy (28 eV), and the rest of the integration time at a lower collision energy (either fixed or ramped between two voltages). This mode ensures that the isobaric tag is sufficiently detached to provide high intensity at low m/z, while also providing sequence information for peptide identification.
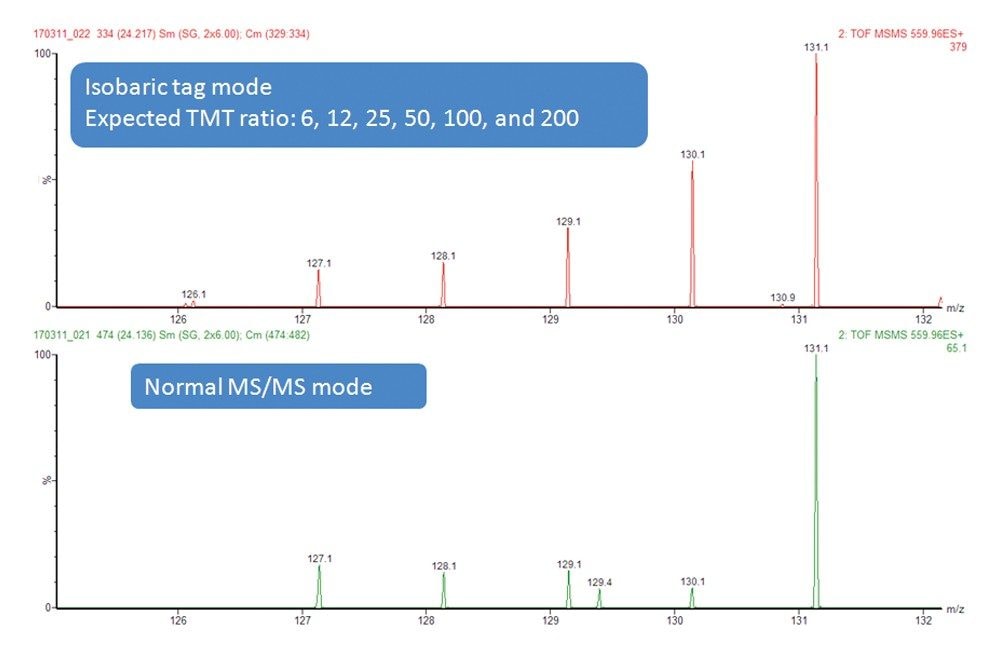
To test the performance of the new algorithm, a complex tryptic digest from the cytosolic fraction of E.coli was labeled with TMT 6.25, 12.5, 25, 50, 100, 200 fmol TMT-labelled BSA digest, and spiked into 1.5 µg of a TMT labelled E.coli digest (1:1:1:1:1:1). The sample was injected and separated on a nanoACQUITY UPLC® System fitted with a microfluidic TRIZAIC UPLC® interface. The NanoTile™ was packed with 1.8 µM ACQUITY UPLC HSS T3 85 µM x 150 mm Column. Peptides were separated using a gradient from 1% to 40% acetonitrile +0.1% formic acid over 90 minutes at a flow rate of 450 nL/min.
The UPLC eluent was passed directly to the nanoflow ion source of a Xevo G2 QTof Mass Spectrometer. For all experiments, 400 ng were injected on column and the new collision energy regime is shown in Figure 1.
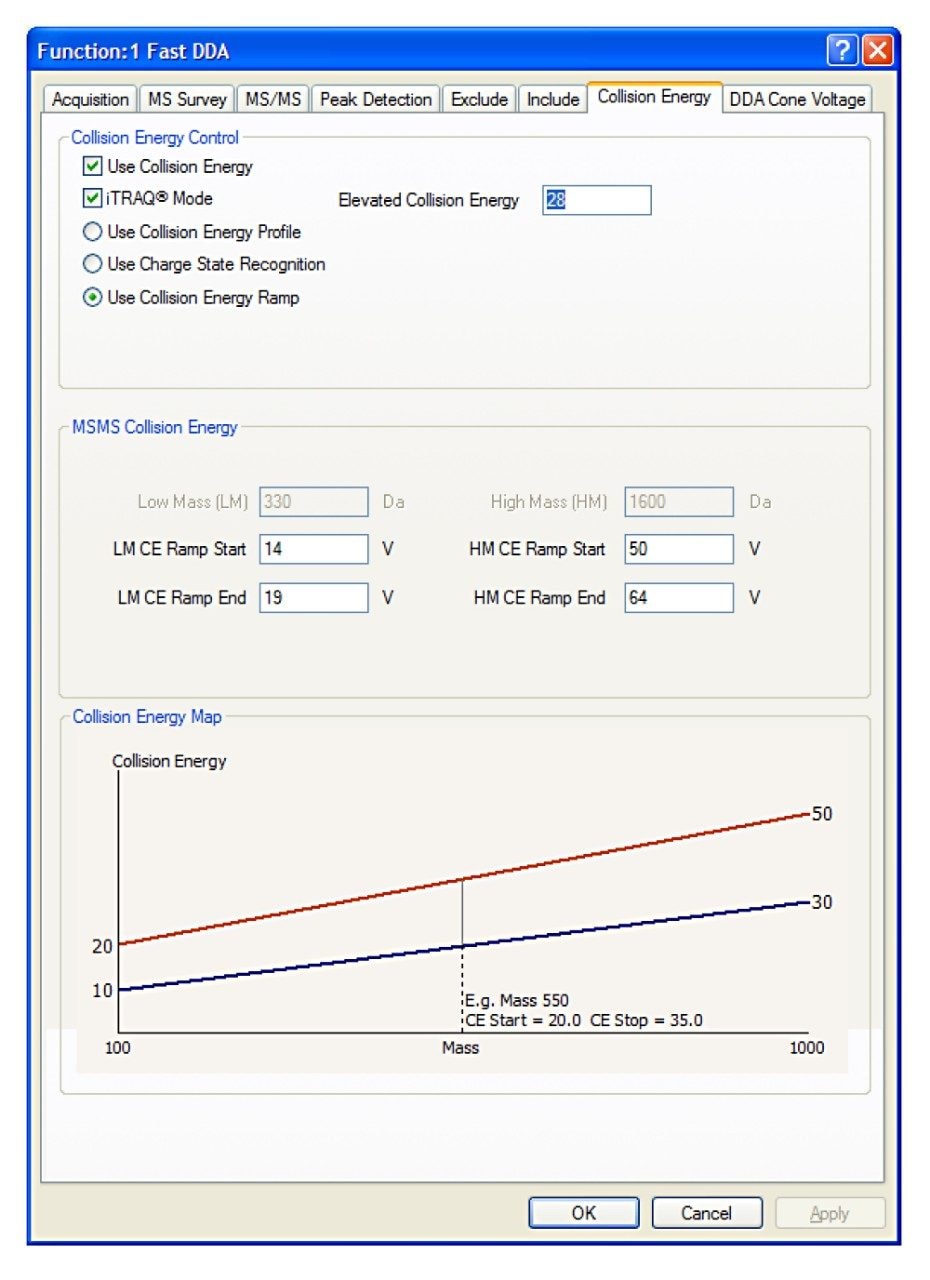
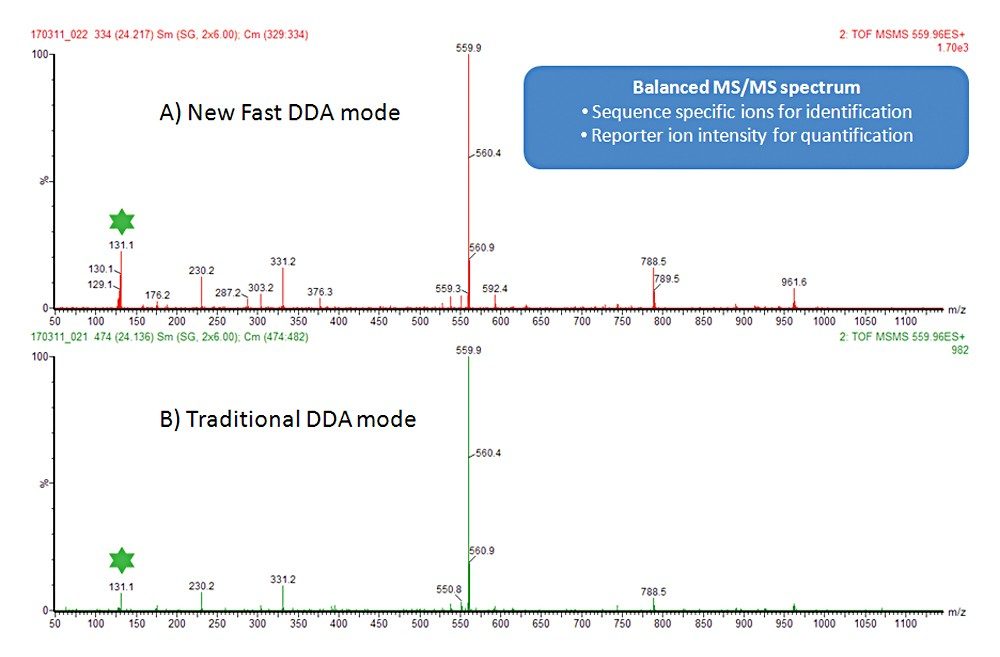
Data were acquired with and without the new isobaric tag mode of operation. In the normal mode a fixed collision energy was used, whereas in the isobaric tag mode, an elevated energy and a lower collision energy were combined (as described above). A comparison of the reporter ion region between the two modes is shown in Figure 2, where the benefits of the new DDA mode for isobaric tag quantification can clearly be seen. The full MS/MS spectrum shown in Figure 3 demonstrates that not just the reporter ion region, but the MS/MS spectrum in general looks well balanced, and contains ions for both quantification and identification purposes. This approach is equally applicable to iTRAQ-labeled samples.
Conclusion
Here we have described a new collision energy regime for the Xevo G2 QTof contained with the FastDDA acquisition mode. The benefit of this mode is that it provides improved results from samples labeled with isobaric tags. These enable more confident results to be obtained for DDA acquisitions in proteomics laboratories for isobaric labeling studies.
Featured Products
720003962, May 2011