This Application note demonstrates UPLC-FLR/MS system as a robust tool for separation and analysis of minor glycoforms or isomers that are otherwise difficult to assign.
Used together, UPLC with detection by FLR and time-of-flight MS comprise a powerful solution for producing required analytical data for batch-to-batch glycan profiling of a recombinant mAb, Trastuzumab. The chromatographic resolution, reproducibility, and mass spectrometry sensitivity enable glycoprofiling of therapeutic antibodies mandated by regulatory agencies. This UPLC-FLR/MS system represents a robust tool for separation and analysis of minor glycoforms or isomers that are otherwise difficult to assign.
Glycosylation plays a vital role in the safety and efficacy of many therapeutic proteins such as recombinant monoclonal antibody (rmAb). Glycosylation of rmAb occurs at the Fc region on the heavy chain (Figure 1). Several studies have shown the correlation between glycosylation variations caused by cell-line selection and changes in culture-medium parameters.1 These variations can have a profound effect on the biological activities of the rmAb drugs, which leads to changes in drug potency in the final product. Regulatory agencies require monitoring of batch-to-batch rmAb drug-production quality, and mandate detailed assessment of the protein glycosylation’s micro-heterogeneity and consistency.
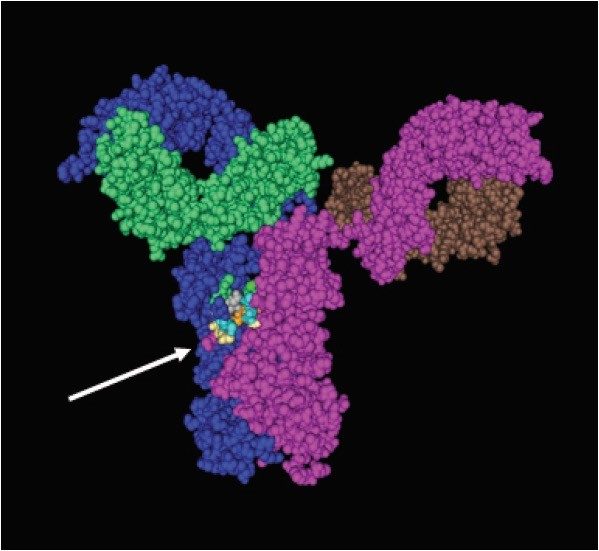
In this study, we applied a robust, sensitive, and reproducible analytical platform that comprises a UltraPerformance LC (UPLC), a fluorescence (FLR) detector, and a XevoTM QTof Mass Spectrometer (MS) for batch-to-batch glycan profiling of an rmAb, Trastuzumab.
Trastuzumab is a therapeutic rmAb (IgG1 subclass) that is widely used for breast cancer treatment. N-linked glycans were released from three batches of Trastuzumab enzymatically, and labeled with a fluorescent tag, 2-aminobenzamide (2-AB). An ACQUITY UPLC HILIC Column was used to separate the released and labeled glycans; the LC was interfaced with the Xevo QTof MS via electrospray ionization. Peak areas from the FLR detector were utilized for glycan quantitation; MS was used for peak assignment using an accurate molecular weight of corresponding glycans.
|
LC system: |
Waters ACQUITY UPLC System |
|
Column: |
ACQUITY UPLC BEH Glycan Column 2.1 x 150 mm, 1.7 μm |
|
Column temp.: |
40 °C |
|
Flow rate: |
400 μL/min |
|
Mobile phase A: |
100 mM ammonium formate, pH 4.5 |
|
Mobile phase B: |
Acetonitrile |
|
Gradient: |
72% to 62% B in 45 min |
|
Weak wash: |
75% acetonitrile |
|
Strong wash: |
20% acetonitrile |
|
Injection: |
5.0 μL partial loop |
|
FLR: |
Waters ACQUITY UPLC Fluorescence Detector |
|
Excitation: |
330 nm |
|
Emission: |
420 nm |
|
Data Rate: |
1 pts/s |
|
PMT Gain: |
1.00 |
|
Time Constant: |
Normal |
|
MS System: |
Waters Xevo QTof MS |
|
Ionization Mode: |
ESI + |
|
Capillary Voltage: |
3200 V |
|
Cone Voltage: |
35 V |
|
Desolvation Temp.: |
350 °C |
|
Desolvation Gas: |
800 L/Hr |
|
Source Temp.: |
120 °C |
|
Acquisition Range: |
800 to 2000 m/z |
|
Collision Energies: |
6 V |
|
Lock Mass: |
Cesium iodide, CSI, (1 μg/μL in 50% isopropanol) |
The rmAb N-linked glycans present in the sample are biantennary and high mannose type. They exhibit considerable heterogeneity and wide dynamic range. Identification and quantification of low-abundant glycans requires sensitive fluorescence detectors. About 5 pmol of sample is typically injected on the column in order to detect minor glycans. The limit of detection for FLR was estimated to lie between 1 to 5 femtomoles.
UPLC HILIC separation provides significantly greater resolution compared to conventional HPLC methods.3 UPLC better resolves isomeric glycans, such as G1 and G1F isomers, makes the data interpretation less ambiguous, and improves quality of quantitation (peak integration).
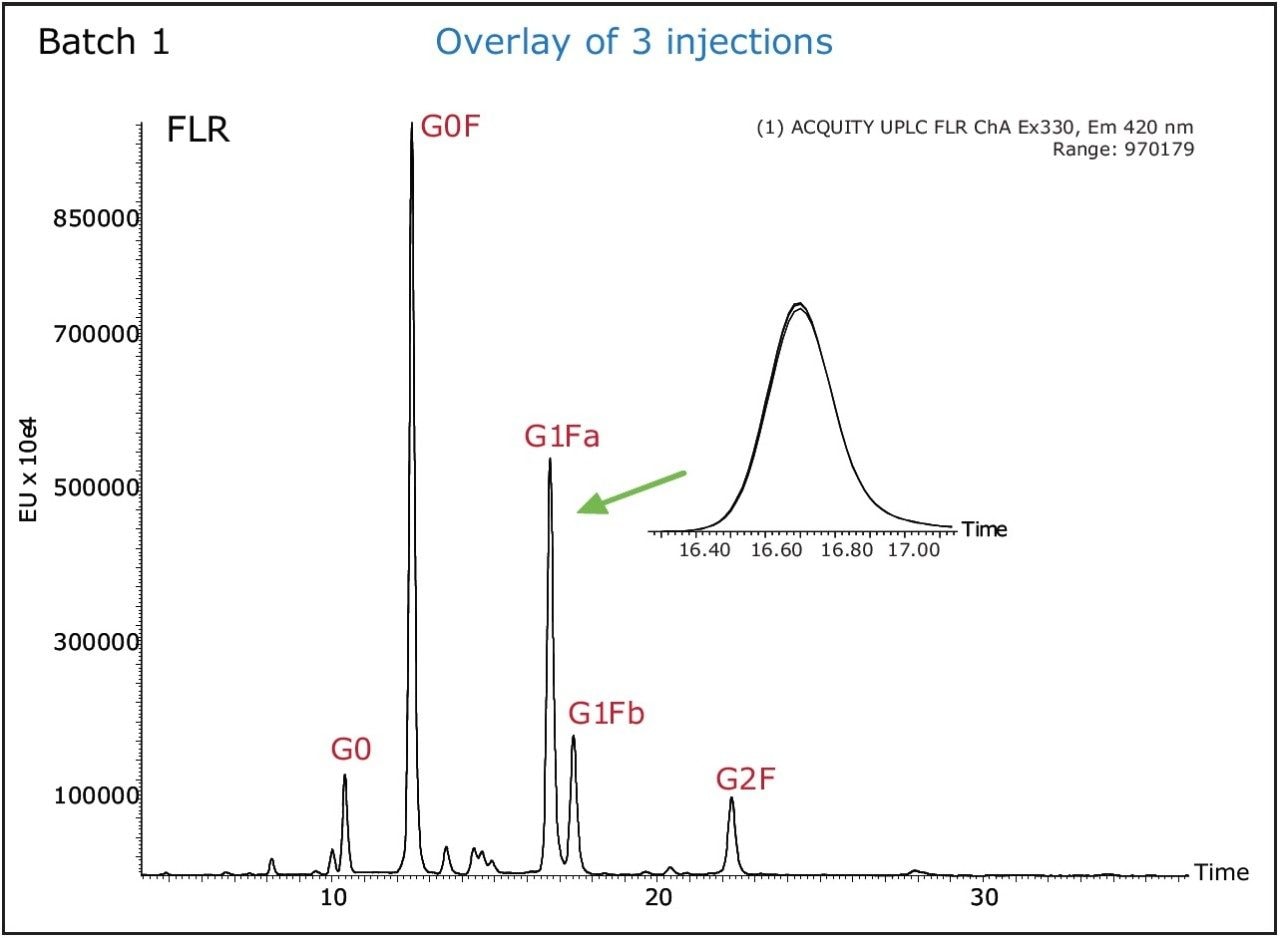
The main purpose of LC/FLR glycans analysis is its relative quantitation. Injection-to-injection variability of UPLC/FLR system was evaluated as shown in Figure 3. The variation (RSD) in peak areas of three injections of the same sample was less than 2% even for minor peaks.
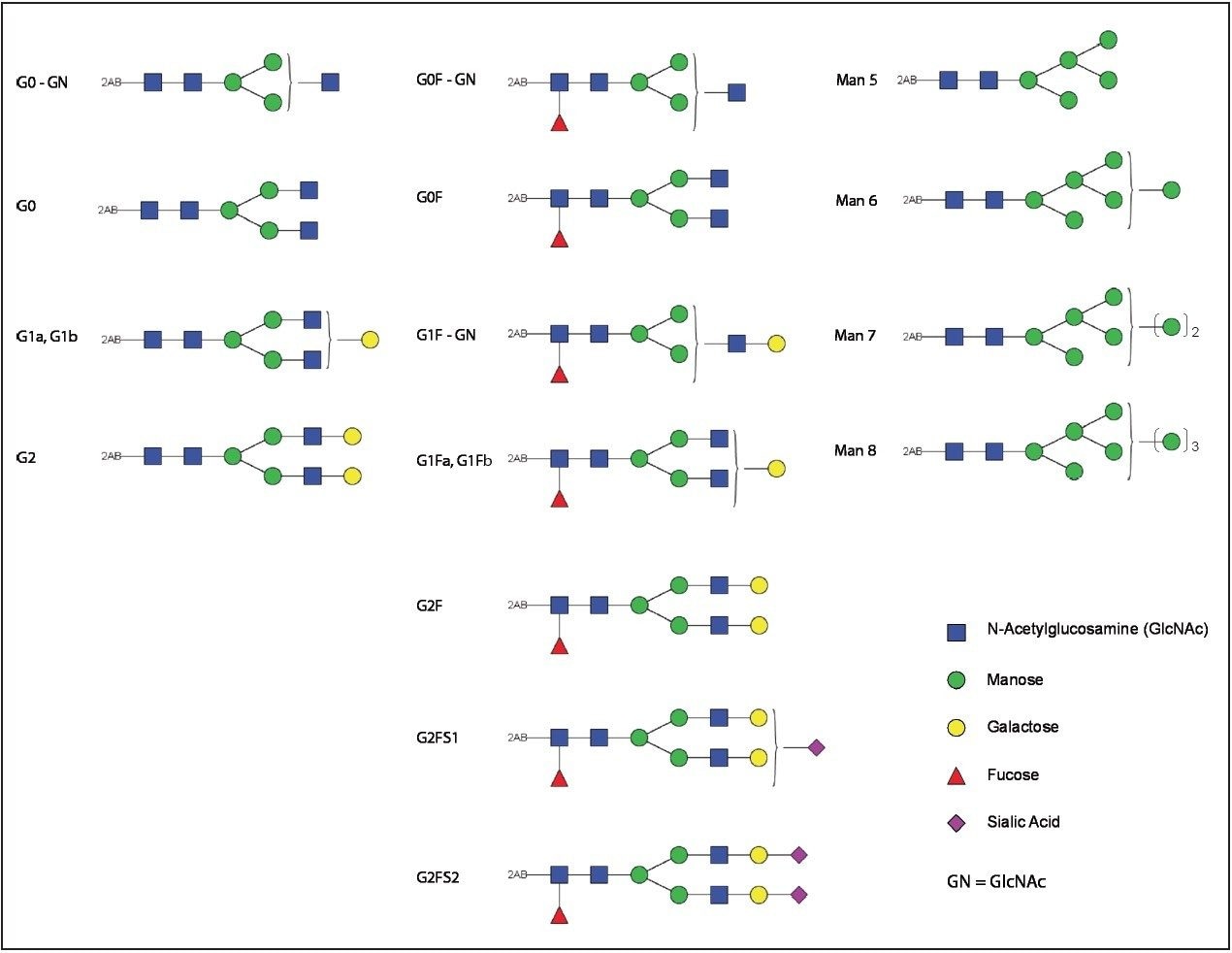
While FLR data are useful for glycan quantitation, MS provides information in addition to chromatographic retention times. Accurate mass data permit the assignment of glycans present in mAb with high confidence (Figures 4A, B). Proposed structures for the 2-AB labeled Trastuzumab glycans are shown in Figure 5.
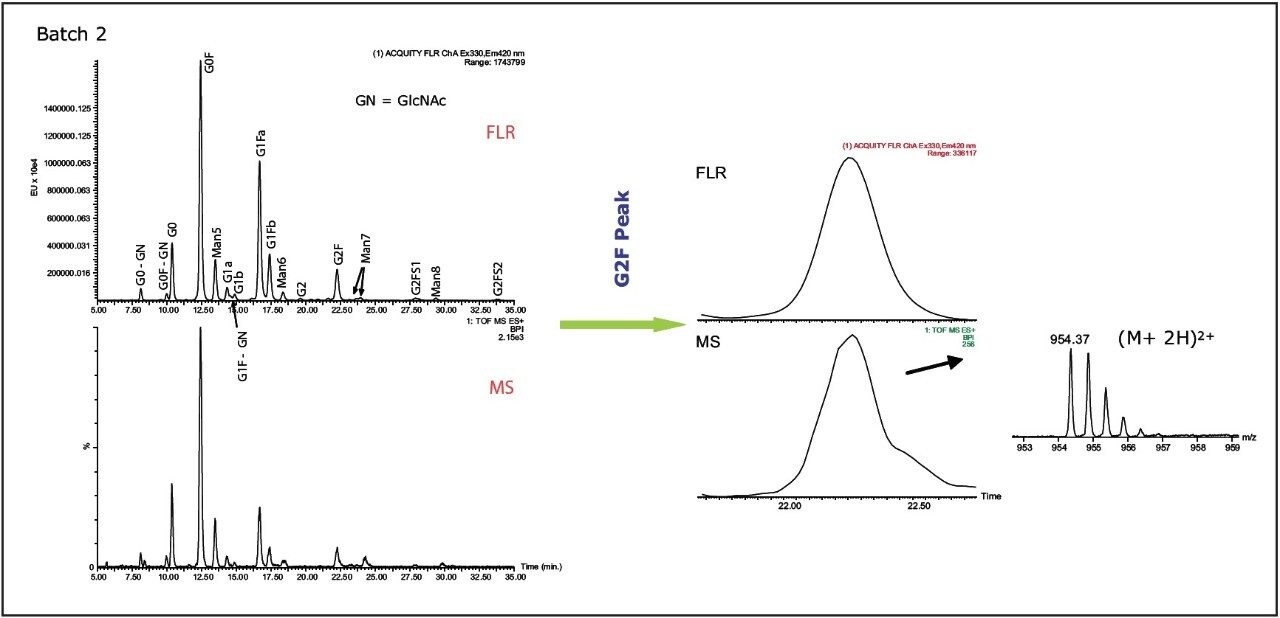
Figure 4A (left). UPLC/FLR/MS analysis of 2-AB labeled glycans from Trastuzumab (Batch 2). The top chromatogram is the FLR chromatogram; the bottom is the MS chromatogram. The glycan identified were confirmed by their accurate mass. Glycan structures are listed in Figure 5.
Figure 4B (right). 2-AB labeled glycan assignment was made by aligning the FLR chromatogram peak with the BPI MS peak. The summed BPI MS scans for G2F peak are shown on the right. The mass error was 20 ppm.
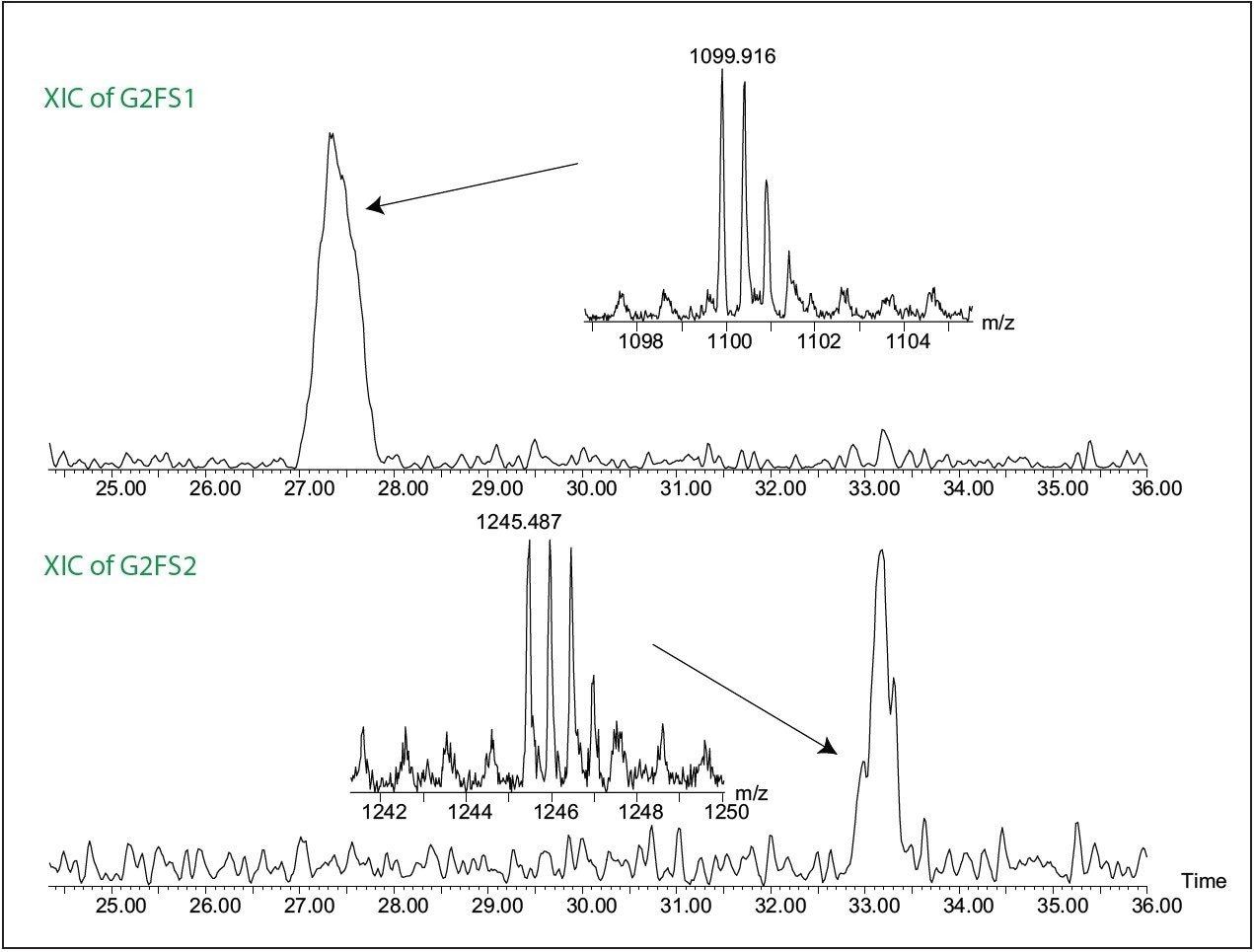
An example of FLR and MS data for G2F peak along with MS spectrum is shown in Figure 3B. MS/MS fragmentation was used to elucidate glycan structure (data not shown). The sensitivity of Xevo QTof MS was sufficient to assign even the minor components. For example, the two sialylated glycans, G2FS1 and G2FS2 with a low fluorescence signal, did not show peaks in the MS base peak ion (BPI) chromatogram. However, the extracted ion chromatograms (XIC) of the doubly charged ions of G2FS1 and G2FS2 glycans clearly confirm the presumed identities (Figure 6).
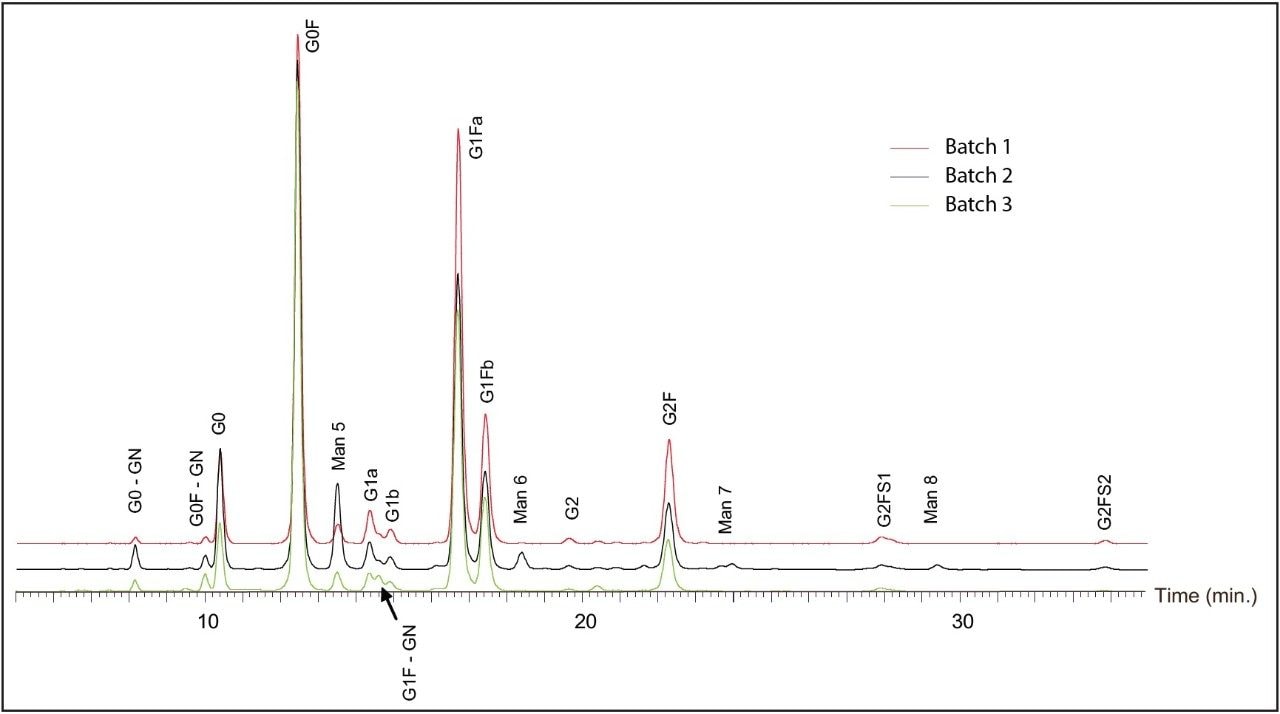
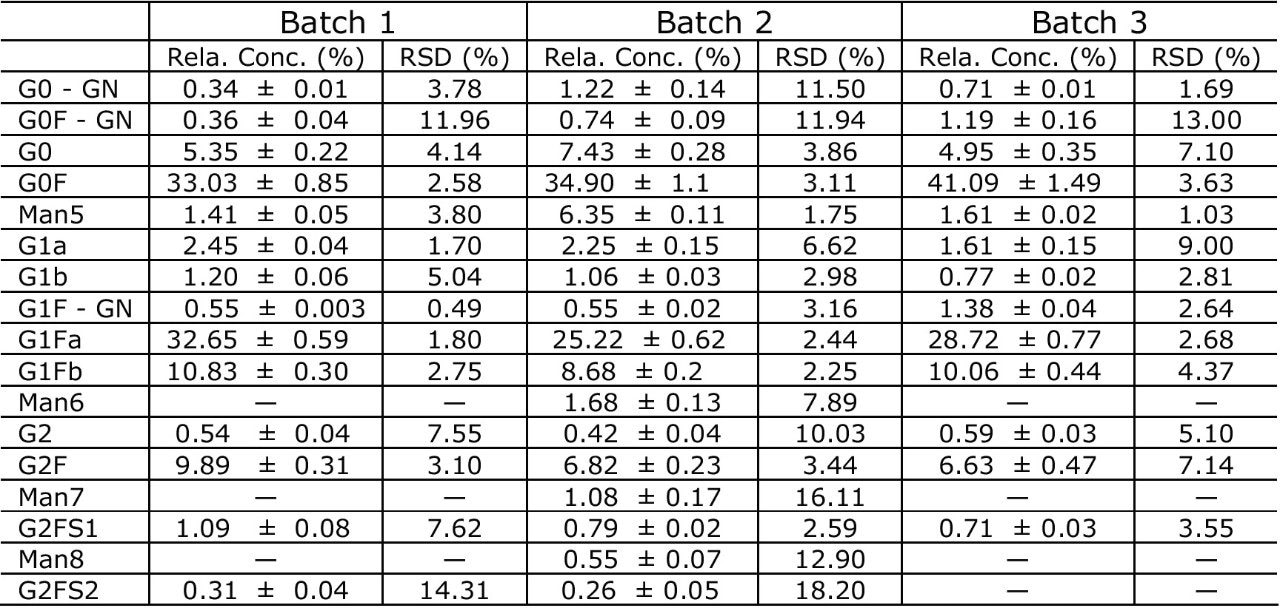
Three different Trastuzumab batches were analyzed (Batch 1 through 3). In order to accurately compare the glycan profiles, the robustness of sample preparation (sample preparation and extraction step using HILIC μElution plate), its variability was also evaluated. Figure 7 shows the overlay of the FLR chromatograms of glycans released from these three Trastuzumab batches, while the graph in Figure 8 compares both the relative glycan abundance and the sample preparation variability. The narrow error bars in confirm that glycan relative quantitation is highly repeatable. The differences between batches of glycan profiles were significantly greater than variability introduced by sample preparation.
Among other differences, we observed significantly higher G0F content in Batch 3 than the other two batches. Man6, Man7, and Man8 were observed only in Batch 2.
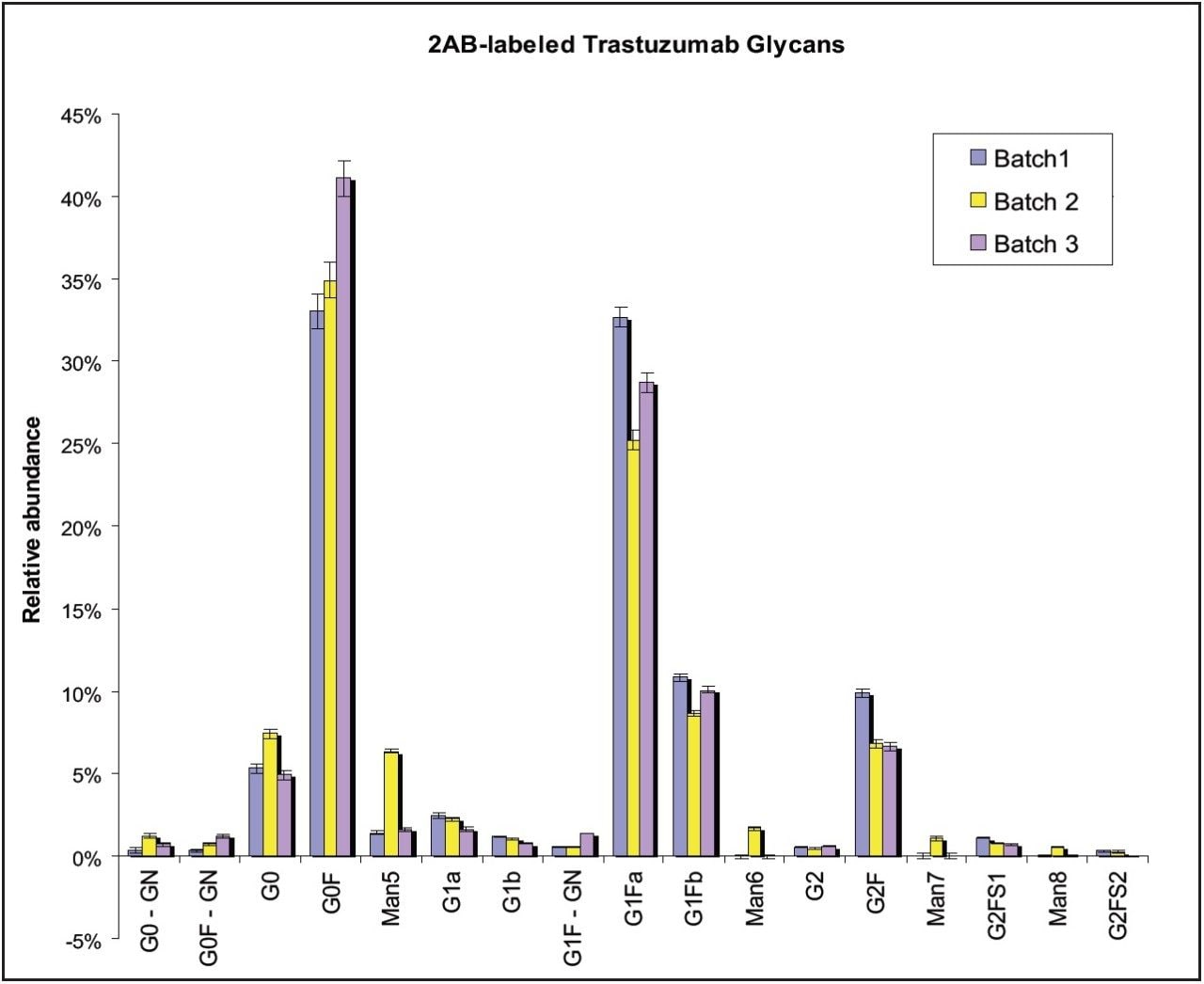
Xevo QTof MS sensitivity was sufficient to confirm the glycan identity for peaks at 0.3% relative intensity (FLR data) of overall glycan content. Table 1 summarizes identified glycans with their relative abundance (%), standard deviation, and %RSD of the integrated FLR peaks (N = 3).
Used together, ACQUITY UPLC with detection by FLR and Xevo QTof MS comprise a powerful system for producing required analytical data. The chromatographic resolution, reproducibility, and mass spectrometry sensitivity enable glycoprofiling of therapeutic antibodies mandated by regulatory agencies. The UPLC-FLR/MS system represents a robust tool for separation and analysis of minor glycoforms or isomers that are otherwise difficult to assign.
This UPLC-FLR/MS platform improves the overall quality of the rmAb-carbohydrate characterization assay and the batch-to-batch consistency test, which are components of drug release tests. The proposed method enables a routine and robust rmAb glycan analysis and may become a tool of choice for biopharmaceuticalrmAb characterization.
720003576, June 2010