In order to ensure public health and safety, reliable analytical methods are necessary to determine pesticide residue levels in foods. Many of the compounds are well suited for gas chromatography (GC) and are often determined in fruits and vegetables using GC with mass spectrometry (GC-MS).
This application note demonstrates an effective cleanup protocol for pesticide analysis in avocado, a highly fatty matrix. After a modified QuEChERS extraction, cleanup is performed using an Oasis PRiME HLB Cartridge. Analysis is performed using GC coupled with atmospheric pressure ionization mass-spectrometry (APGC-MS/MS).
In recent years, food safety laboratories have adopted new and simplified sample preparation methods designed to reduce analysis time and related costs, as well as to increase throughput. For example, the QuEChERS methods for fruits and vegetables require only minutes for sample preparation and replace prior methods that took hours or days. In this study, this type of simplified sample preparation is applied to pesticide analysis in avocado, a fruit matrix of very high lipid content. A typical avocado contains 10–15% fat and about 1% total phospholipids. In the QuEChERS extraction, significant amounts of the fat and phospholipids are co-extracted along with the target pesticides. The presence of these co-extracted substances, particularly the phospholipids, can lead to chromatographic interference, contamination of the GC injector and column, and contamination of the mass spectrometer itself. To avoid these complications, a cleanup step is recommended prior to the instrumental analysis. This is typically performed using dispersive SPE with mixed sorbents, often with cumbersome multi-step centrifugation. In this study an Oasis PRiME HLB Cartridge was used for a simple pass-through cleanup to effectively remove fats and phospholipids. This method was applied to a number of pesticides registered for use on avocado in various world markets and suitable for GC-MS analysis. The APGC methodology was used for quantitative analysis in this study (APGC-MS/MS).
|
GC system: |
Agilent 7890 |
|
Column: |
Restek Rxi-5 ms, 30 m x 0.25 mm x 0.25 μm |
|
Flow rate: |
1.0 mL/min Helium |
|
Injection volume: |
1 μL (15:1 split) |
|
Temperature program: |
80 °C initial, hold for 0.5 min, 12 °C/min to 320 °C and hold for 8 min |
|
Mass spectrometer: |
Xevo TQ-S |
|
Ion mode: |
API+ (charge transfer mode) |
|
Corona: |
2.8 μA |
|
Source temperature: |
150 °C |
|
Probe temperature: |
450 °C |
|
Cone gas: |
170 L/Hr |
|
Auxiliary gas: |
250 L/Hr |
|
Collision gas: |
0.15 mL/min (Ar) |
|
Nebulizer: |
4.0 bar |
|
Data management: |
MassLynx v4.1 |
|
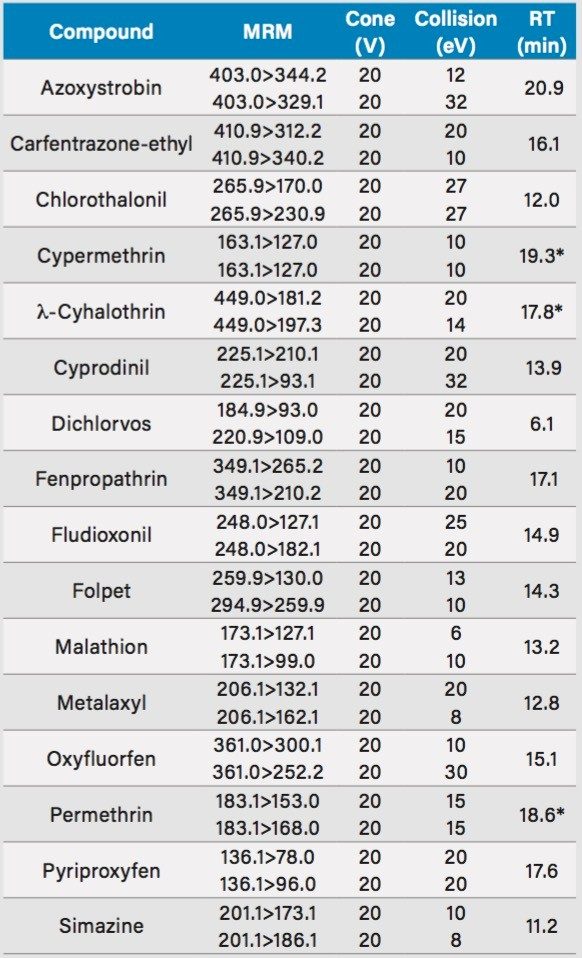
Other instrument parameters are presented in Table 1. |
AOAC QuEChERS Extraction: Avocado is so high in fat, the AOAC QuEChERS method is modified to reduce the sample size from 15 g to 5 g. Weigh 5 g sample into a 50 mL centrifuge tube (for a spiked sample, add the required volume of spiking standard solution). Add 5 mL water and 15 mL 99:1 acetonitrile/acetic acid. Vortex for 30 seconds and shake well for 2 minutes. Add QuEChERS salts (contents of DisQuE pouch for AOAC, p/n 186006812). Shake the tube vigorously by hand for 1 minute and centrifuge at approximately 2500 rcf for 5 minutes. An aliquot of the supernatant extract (top layer) is taken for analysis.
Pass-through SPE Cleanup: Install an Oasis PRiME HLB Cartridge (3 cc, 60 mg, p/n 186008056 ) on a vacuum manifold. Set to minimal vacuum (~2 in Hg). Pass 0.4 mL of the QuEChERS extract through the cartridge to waste. Install collection vessels. Pass 0.6 mL of the QuEChERS extract and collect. In this study for APGC-MS analysis 200 µL of the collected extract is transferred to a Qsert Vial and analyzed directly. Alternatively, a portion of the collected extract can be evaporated and reconstituted in toluene for splitless GC injection.
It is important to distinguish any recovery losses resulting from the SPE cleanup from losses resulting from the initial QuEChERS extraction. Therefore, the modified QuEChERS procedure was evaluated for recovery of the target compounds prior to any SPE recovery experiments. All compounds (spiked at 40 ng/g) were recovered at greater than 80% with the exception of folpet (70%), fenpropathrin (65%), and pyriproxyfen (75%).
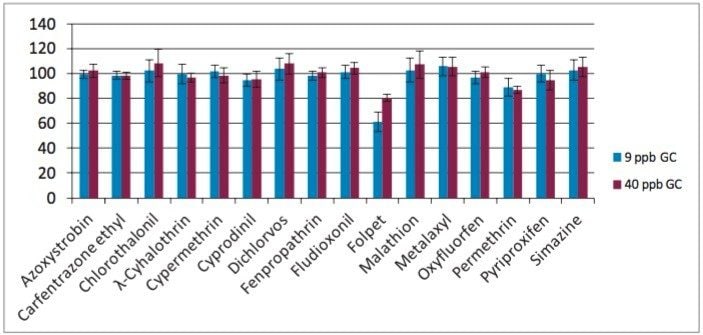
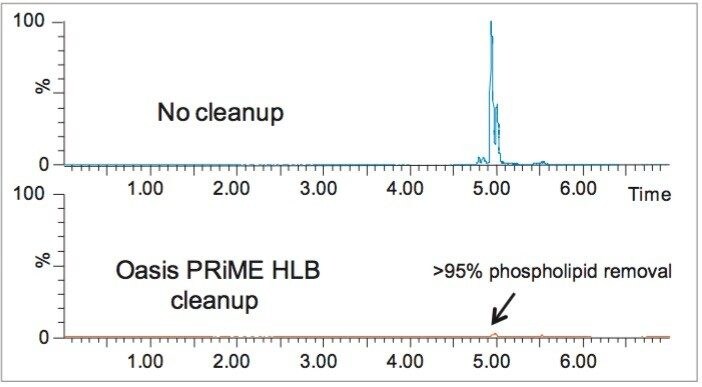
SPE cleanup recovery data (see Figure 1) were determined using blank avocado samples obtained using the modified QuEChERS protocol. Blank extracts were spiked at the 9 and 40 µg/kg (ppb) levels and were subjected to the pass-through SPE cleanup protocol. Response for each compound was compared with response obtained from identical blank sample extracts spiked after the SPE cleanup. Only folpet, a thermal and pH labile substance, showed recovery losses greater than 20% resulting from the cleanup protocol. Figure 2 shows that the Oasis PRiME HLB Cartridge cleanup removed greater than 95% of phospholipids from the avocado QuEChERS extract. Also, greater than 90% of chlorophyll and approximately 80% of fat was removed in the cleanup. Cleanup obtained in only seconds with this protocol was comparable to traditional dispersive SPE (dSPE) cleanups that often require multiple cumbersome centrifugation steps.
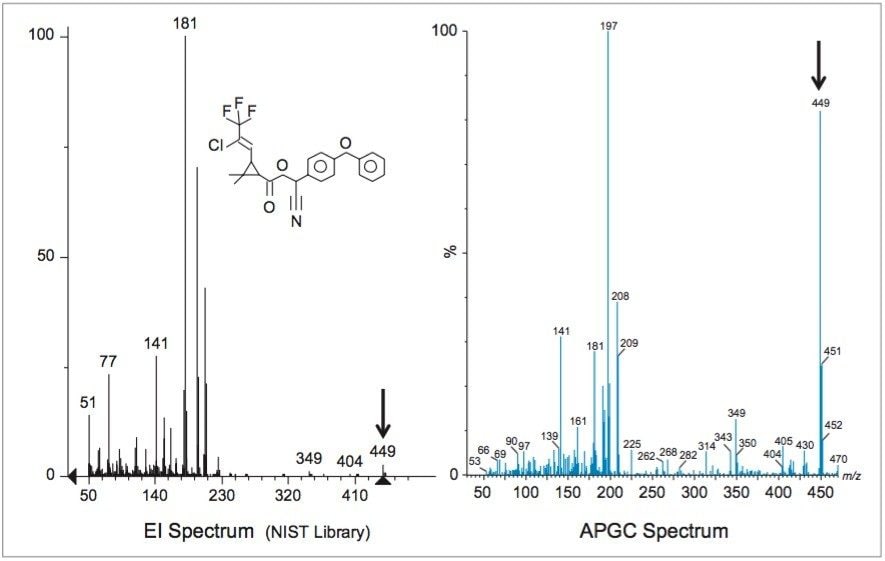
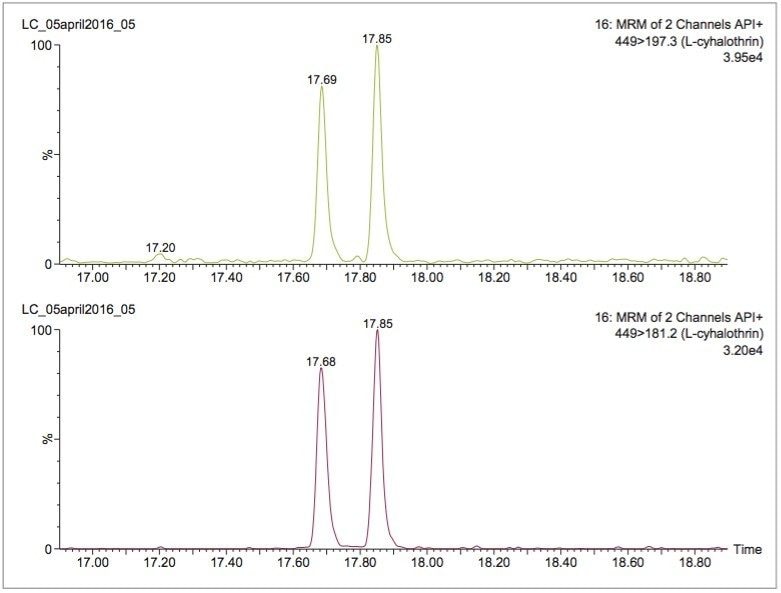
Compared with traditional electron-impact mass spectrometry (EI-MS), the API ionization used for APGC is a softer form of ionization and is often a superior technique for tandem MS. The softer ionization results in less in-source fragmentation and a greater likelihood of obtaining a molecular ion for subsequent fragmentation in the collision cell. An example is shown in Figure 3, a comparison of the API+ and EI+ mass spectra obtained for λ-cyhalothrin. Note the high abundance of the molecular ion (m/z 449) in the API spectrum compared with the EI spectrum. In this study, two MRM transitions were monitored for determination of λ-cyhalothrin (see Table 1). Each of these transitions resulted from fragmentation of the m/z 449 molecular ion; these transitions would not be possible using EI-MS. Figure 4 shows the APGC-MS/MS determination of the isomers of λ-cyhalothrin spiked at 9 ng/g in avocado after QuEChERS extraction and Oasis PRiME HLB cleanup. Similar low ng/g (ppb) detection limits were observed for all the target compounds in this study.
720005816, October 2016