
Perfluoroalkyl substances (PFASs) encompass a range of fully fluorinated alkyl compounds, typically with an anionic end group. These compounds have been implemented in a range of consumer goods and industrial processes due to their hydro- and lipo-phobic properties. As a result of their widespread use and subsequent leaching from materials, they have been found in various environmental and biological samples.
Concern that these compounds exhibit characteristics of persistent organic pollutants (POPs) has resulted in study and legislation against their use. For monitoring and research purposes, sub-ppb detection of these compounds is often required. Traditionally, this type of analysis has been performed using the selective MRM approach on a tandem quadrupole MS. However, the ability to identify other contaminants of concern post acquisition or matrix components such as co-extracted bile acids supports the use of high resolution mass spectrometry (HRMS).
HRMS screening techniques can, in theory, monitor an unlimited number of targets at the same time as providing information to help discover unknown compounds or metabolites of interest. The ease of use and efficacy of a non-targeted, data independent, analysis type (MSE), coupled with a state-of-the-art scientific information system (UNIFI) for multi-analyte screening in environmental samples is demonstrated.
This approach was undertaken on an authentic sample analysis of mink liver for low level detection (ppb and sub-ppb) and quantification of identified PFASs. This particular application note will focus on introducing the novel way a user in a routine environment can customize data review within the scientific information system to establish an fast, concise, and consistent approach to reviewing HRMS data.
Reviewing complex high resolution, non-targeted MSE or HDMSE datasets using workflows, filters, and views within an integrated scientific information system allows:
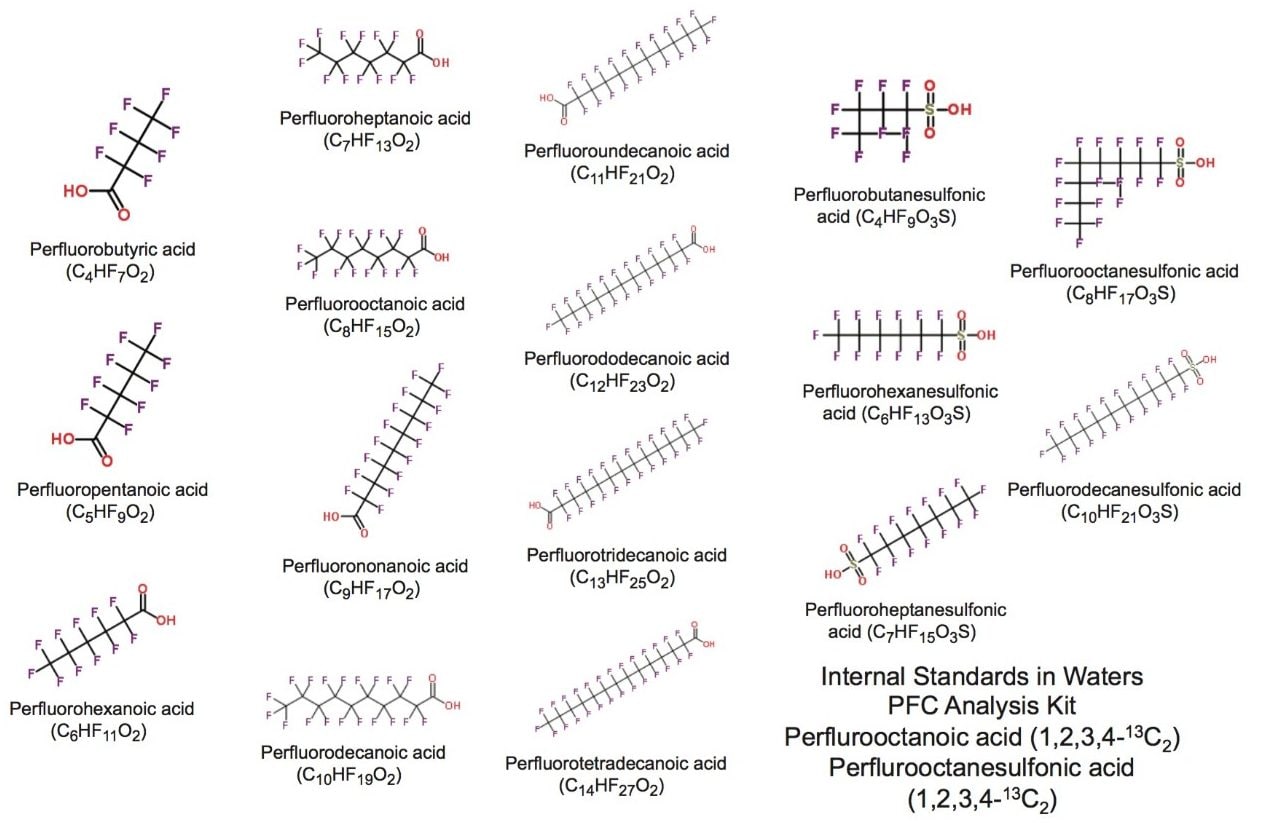
Perfluoroalkyl substances (PFASs) encompass a range of fully fluorinated alkyl compounds, typically with an anionic end group (Figure 1). These compounds have been implemented in a range of consumer goods and industrial processes due to their hydro- and lipo-phobic properties. As a result of their widespread use and subsequent leaching from materials, they have been found in various environmental and biological samples. Concern that these compounds exhibit characteristics of persistent organic pollutants (POPs) has resulted in study and legislation against their use.1 For monitoring and research purposes, sub-ppb detection of these compounds is often required. Traditionally, this type of analysis has been performed using the selective MRM approach on a tandem quadrupole MS. However, the ability to identify other contaminants of concern post acquisition or matrix components such as co-extracted bile acids2 supports the use of high resolution mass spectrometry (HRMS).
High resolution mass spectrometry (HRMS) screening techniques can, in theory, monitor an unlimited number of targets at the same time as providing information to help discover unknown compounds or metabolites of interest. The ease of use and efficacy of a non-targeted, data independent, analysis type (MSE),3 coupled with a state-of-the-art scientific information system (UNIFI) for multi-analyte screening in environmental samples is demonstrated with this case study. This approach was undertaken on an authentic sample analysis of mink liver for low level detection (ppb and sub-ppb) and quantification of identified PFASs. This particular application note will focus on introducing the novel way a user in a routine environment can customize data review within the scientific information system to establish a concise, rapid, facile, and consistent approach to reviewing HRMS data.
|
LC system: |
ACQUITY UPLC I-Class with the PFC Analysis Kit |
||
|
Column: |
ACQUITY UPLC BEH C18, 1.7 μm 2.1 x 50 mm |
||
|
Column temp.: |
55 °C |
||
|
Mobile phase A: |
98:2 Water: MeOH 2 mM ammonium acetate |
||
|
Mobile phase B: |
MeOH 2 mM ammonium acetate |
|
Time (min) |
Flow rate (mL/min) |
%A |
%B |
|---|---|---|---|
|
Initial |
0.65 |
90 |
10 |
|
0.5 |
0.65 |
90 |
10 |
|
5.1 |
0.65 |
0 |
100 |
|
6.6 |
0.65 |
0 |
100 |
|
6.7 |
0.65 |
90 |
10 |
|
8.5 |
0.65 |
90 |
10 |
|
MS system: |
Xevo G2-XS QTof |
||
|
Acquisition range: |
50 to 1200 m/z |
||
|
Ionization mode: |
ESI |
||
|
Capillary voltage: |
1.5 kV |
||
|
Cone voltage: |
15 V |
||
|
Source temp.: |
120 °C |
||
|
Desolvation temp.: |
550 °C |
||
|
Cone gas flow: |
50 L/hr |
||
|
Desolvation gas flow: |
1000 L/hr min. |
||
|
Scan time: |
0.2 s |
||
|
Low collision energy: |
6 V |
||
|
High collision energy: |
35 to 75 V |
||
|
Lock mass: |
Leucine enkephalin (554.2610) |
Solvent standards of the PFASs investigated were diluted in methanol from a mixed standard provided in Waters PFC Analysis Kit (p/n 176001744). Mink liver samples were provided by a collaborator as extracts, per the methodology described in Kärrman et. al..4 Mink liver samples were stored in vials prior to analysis, and were diluted 1:10 in methanol before injection onto the system.
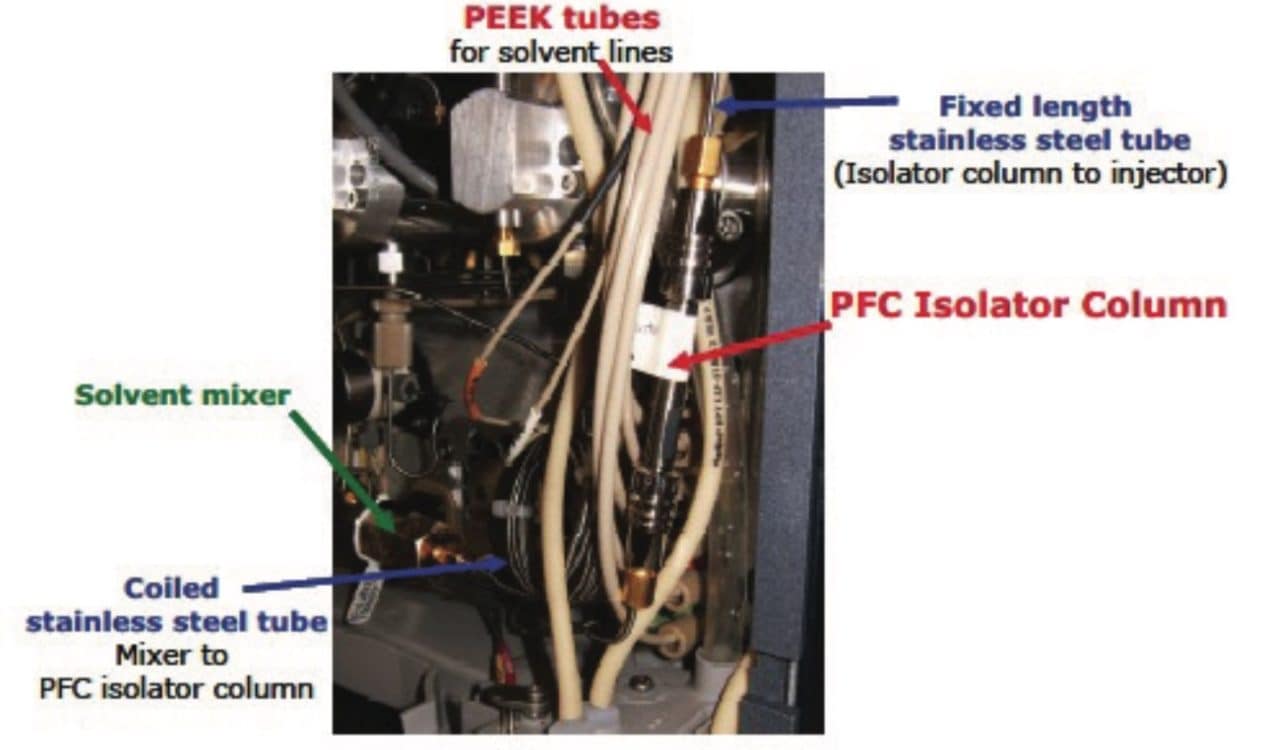
In order to ensure that possible sources of PFASs contamination present within all LC analytical equipment would not bias the analysis, modifications to the UPLC System were made. Using components from the PFC Analysis Kit, Teflon (a common source of PFAS contamination) solvent tubing was replaced with PEEK tubing provided in the kit. A 50-mm ACQUITY UPLC BEH C18 Isolator Column was also placed after the solvent mixing mechanism, and prior to the sample manager, to retain any residual PFASs that were present in the system or solvents. More information regarding the PFC Analysis Kit can be found in Waters Application Note no. 720002813en.5 Figure 2 shows the placement of the BEH C18 Isolator Column.
Standards and samples were analyzed using the ACQUITY UPLC I-Class System coupled to the Xevo G2-XS QTof Mass Spectrometer. A non-targeted, data independent analysis, (MSE)1 was collected and processed in the UNIFI Scientific Information System.
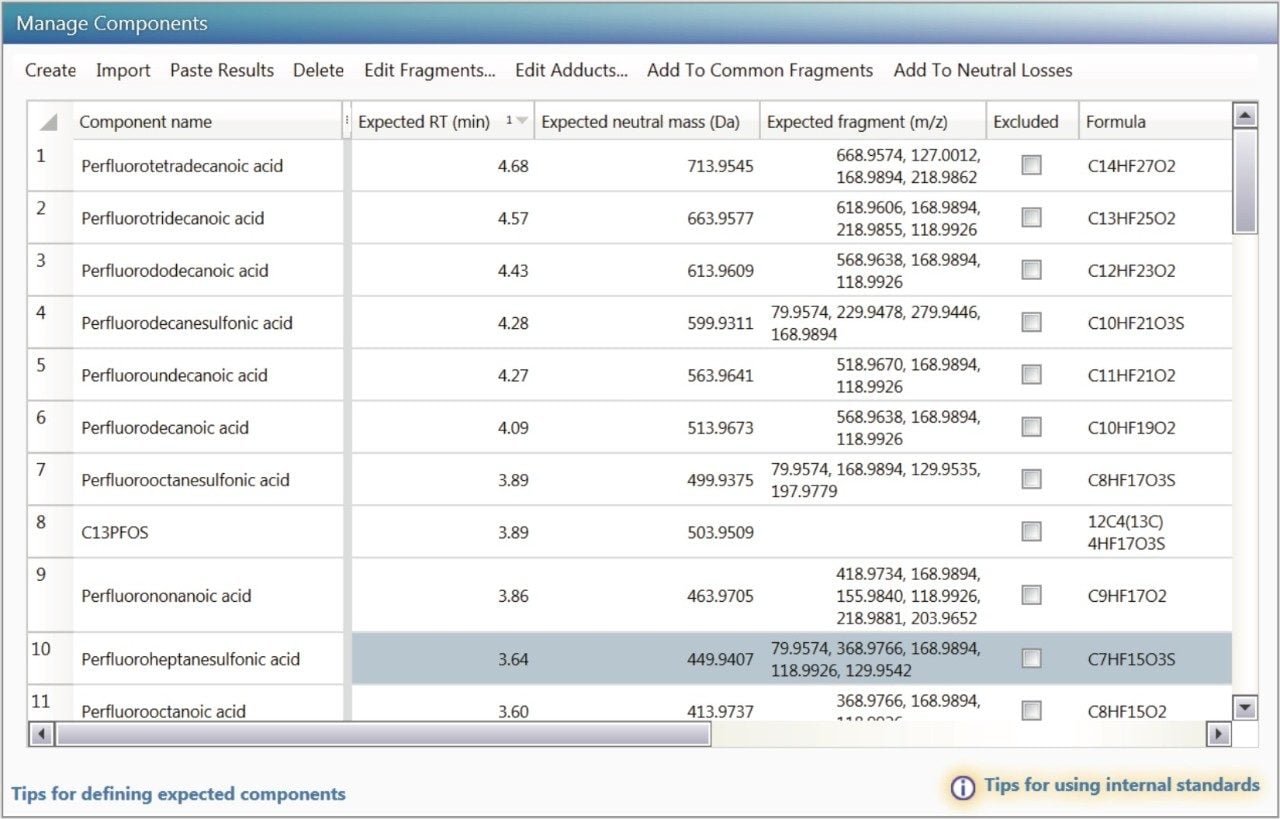
The aim of these case studies is to show how a user can get from injection of a sample to submission of an accurate report in a quick, efficient, systematic, and reproducible way using the workflows, views filters, and software tools in UNIFI.6 Here, the component list provided a flexible and information rich approach to making identifications in the analyzed samples. The component list displayed in Figure 4 contains detection results such as retention time and theoretical accurate mass product ions. It can be updated at any time following data acquisition, thus affording historical data review for emerging compounds that become of interest following an original analysis. UNIFI’s Scientific Library functionality makes this informative updating of screening lists possible, and is described later in this application note.
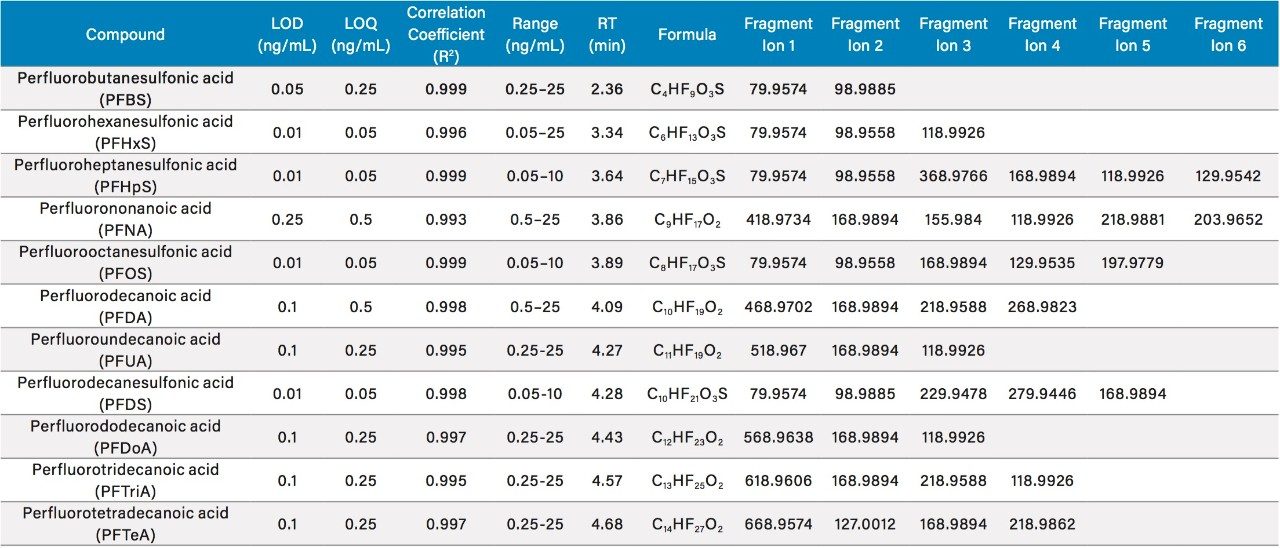
For the 11 carboxylic and sulfonic acid PFASs, instrumental performance with respect to determination of limits of detection (LOD; peak-to-peak S/N 1:3), quantification (LOQ; peak-to-peak S/N 1:10), and linear dynamic range were carried out with solvent standards that are listed in Table 1. Theoretical exact mass product ion information was also obtained, where at least two ions were observed for each compound.
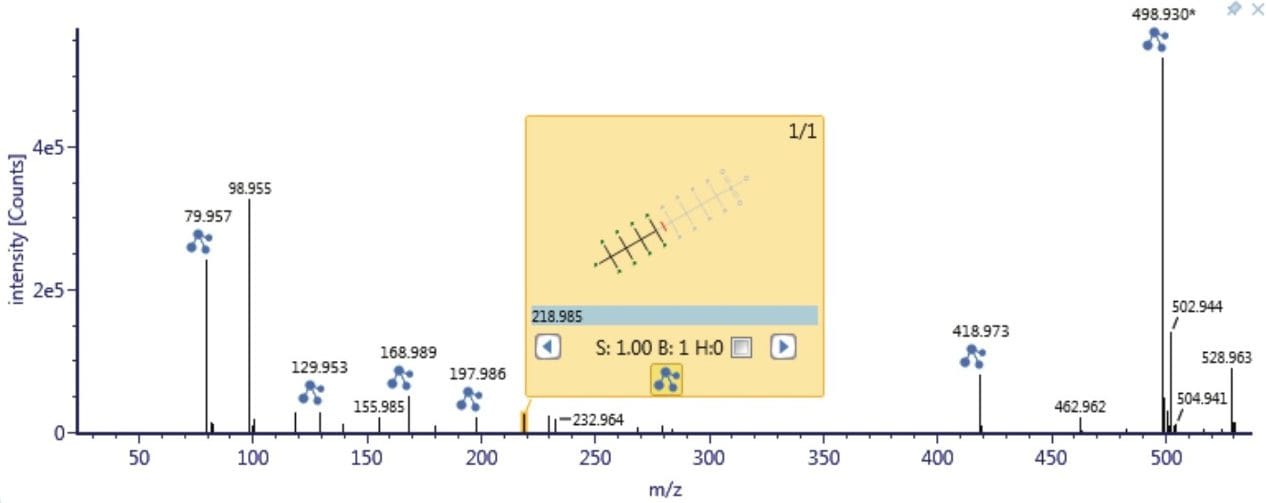
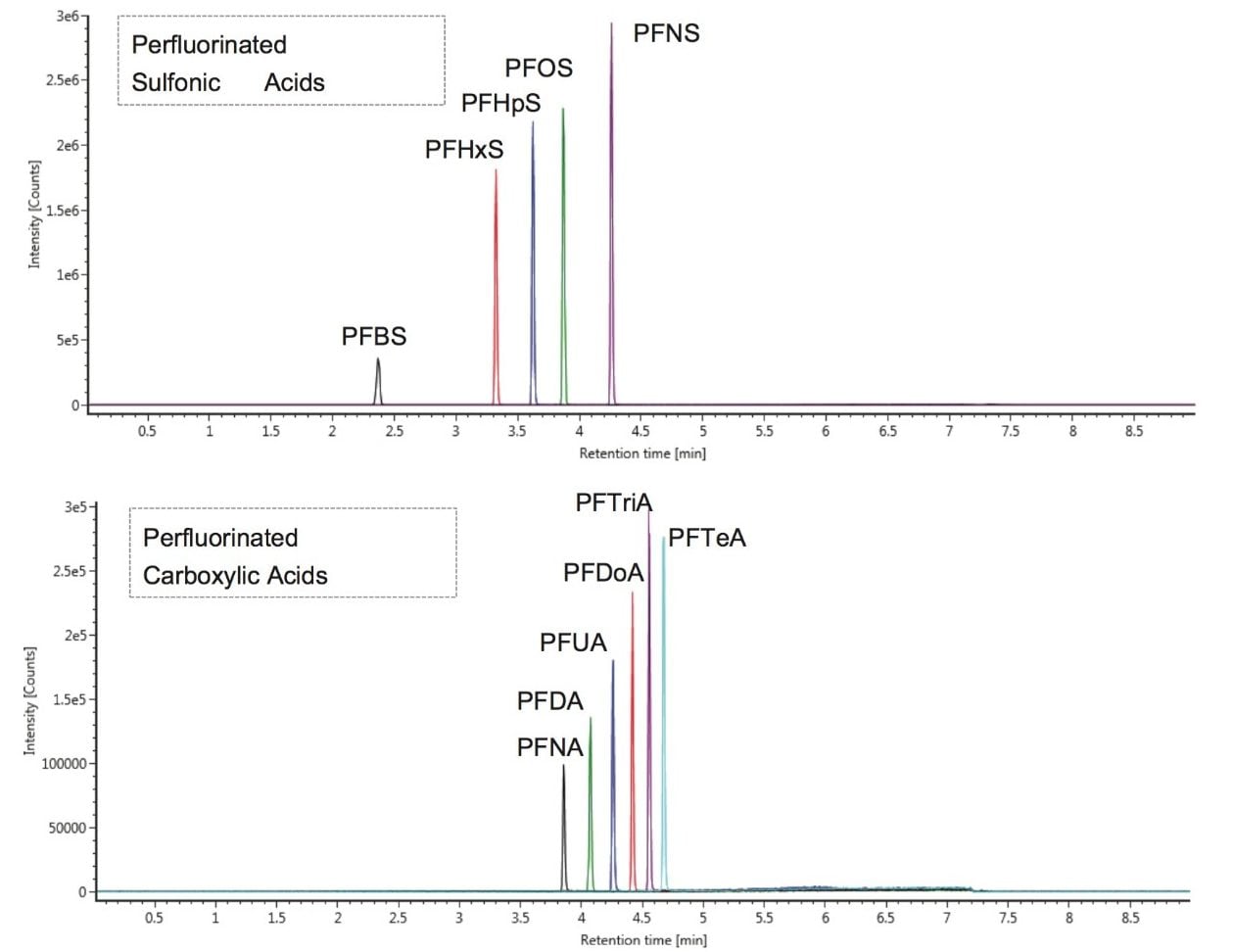
As a result of full spectral acquisition, the entirety of the fragmentation pathways of the analytes of interest is obtained, as is shown in Figure 5. Chromatographic separation of the PFASs standards analyzed is shown in Figure 6. Peaks were approximately 6 s in width at the base, and between 12 to 15 points across the peak. This was maintained for both low and high energy data, due to the sufficient scan speeds of the Xevo G2-XS QTof instrument.
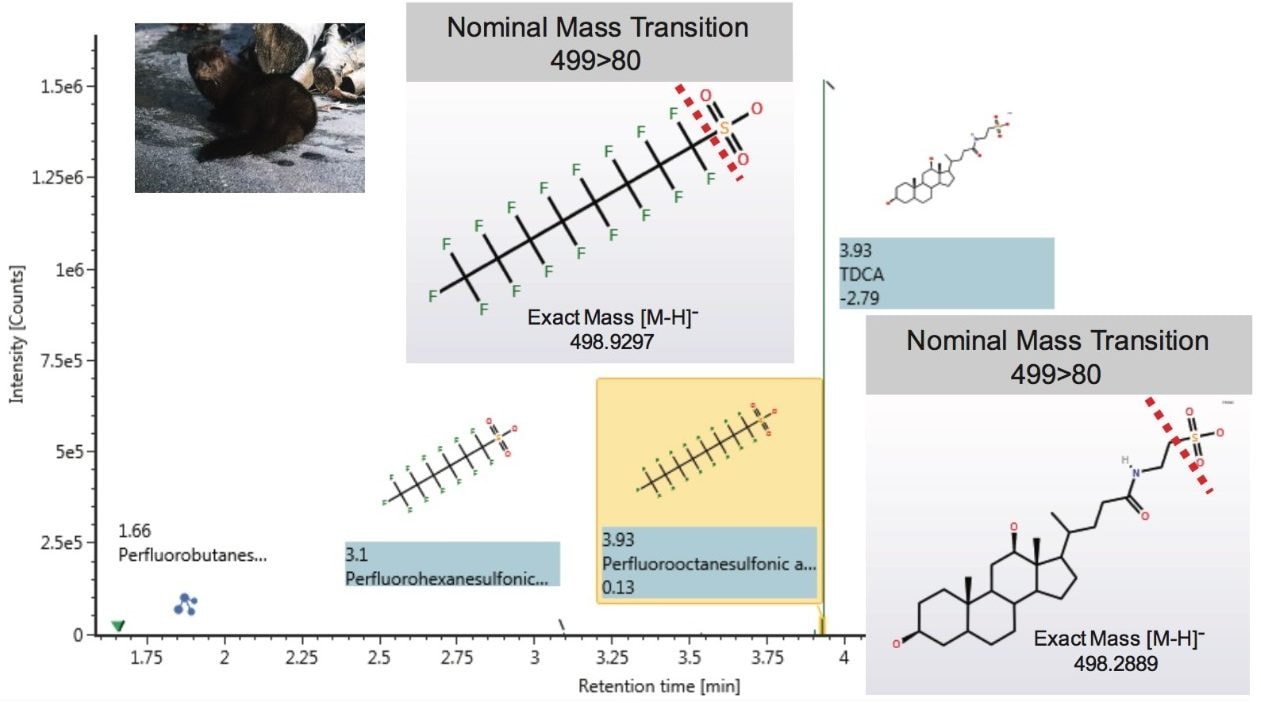
Diluted mink liver extracts contained various PFASs including PFHpS, PFHxS, PFBS, PFOS, PFDS, and PFNA that were detected at levels ranging from 0.2 to 0.8 ng/mL (without dilution and sample mass correction). In addition to isolating the PFASs of interest, the extraction method also resulted in the co-extraction of the bile acid taurodeoxcyholate (TDCA), which co-elutes chromatographically with PFOS. This can result in a high intensity peak in the analysis. However using exact mass measurements afforded by QTof MS, TDCA can be easily distinguished from PFOS (Figure 7).
The Xevo G2-XS QTof is a highly sensitive high resolution mass spectrometer able to detect carboxylic and sulfonic PFASs at sub-ppb levels with exact mass measurements for both precursor and product ions.
Exact mass fragment information obtained in the same analysis for any analyte of interest can easily be compared with the parent structure to provide specific and comprehensive structural information.
Data independent full spectral acquisition allows the user to see various unexpected or unintended aspects of a sample, as well as the ability to perform historical data review.
Drs. Anna Rotander and Ingrid Ericson Jogsten of the MTM Research Centre are kindly acknowledged for providing the mink liver extracts.
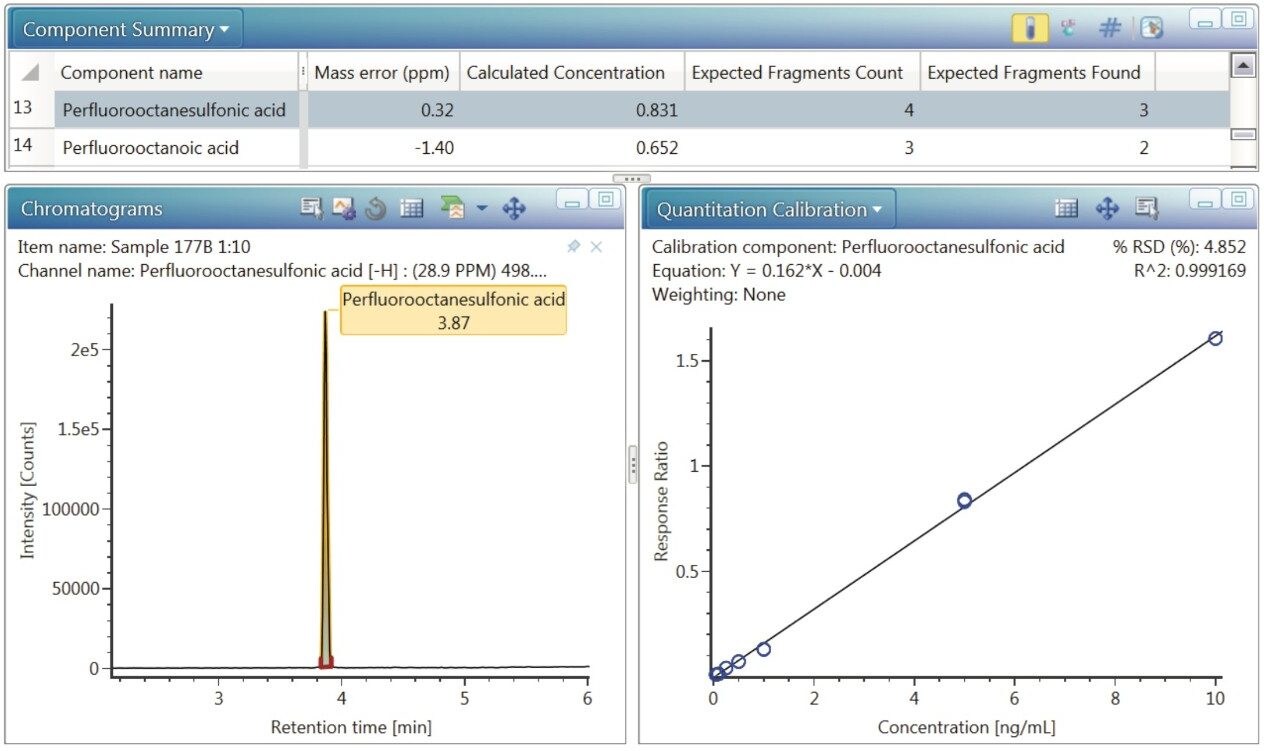
Following qualification of PFASs in the mink liver samples, quantification was performed for all identified analytes present above the LOQs and within the linear dynamic range. A user created workflow step (Figure 8) was created to show relevant displays such as a calibration curve (which includes the R2 value, slope calculation, and weighting used, as well as %RSD value), calculated concentration, identified and expected high energy product ions, and an extracted ion chromatogram for the identified analyte – all with a single mouse click. The workflow step is applied to the samples only, which is also user defined when designing the workflow step. Figure 9 shows these results for PFOS in mink liver.
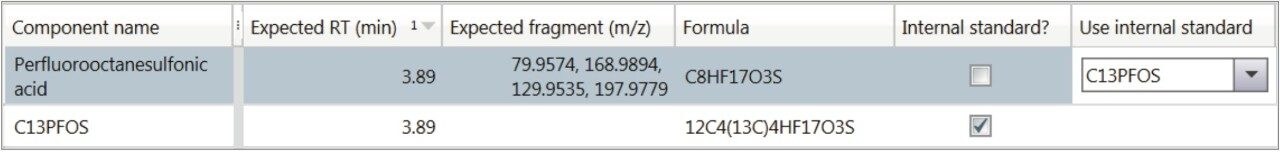
Internal standards (13CPFOS and 13CPFOA) were used to correct for any variations in injection or standard conditions. Their application to the quantitative analysis are input in the analysis method target list, and coupled to the analytes through a drop down menu. This is shown in Figure 10. This information is input prior to injection, such that all quantitative and qualitative analyses performed occur during the live injection time, within UNIFI Software.
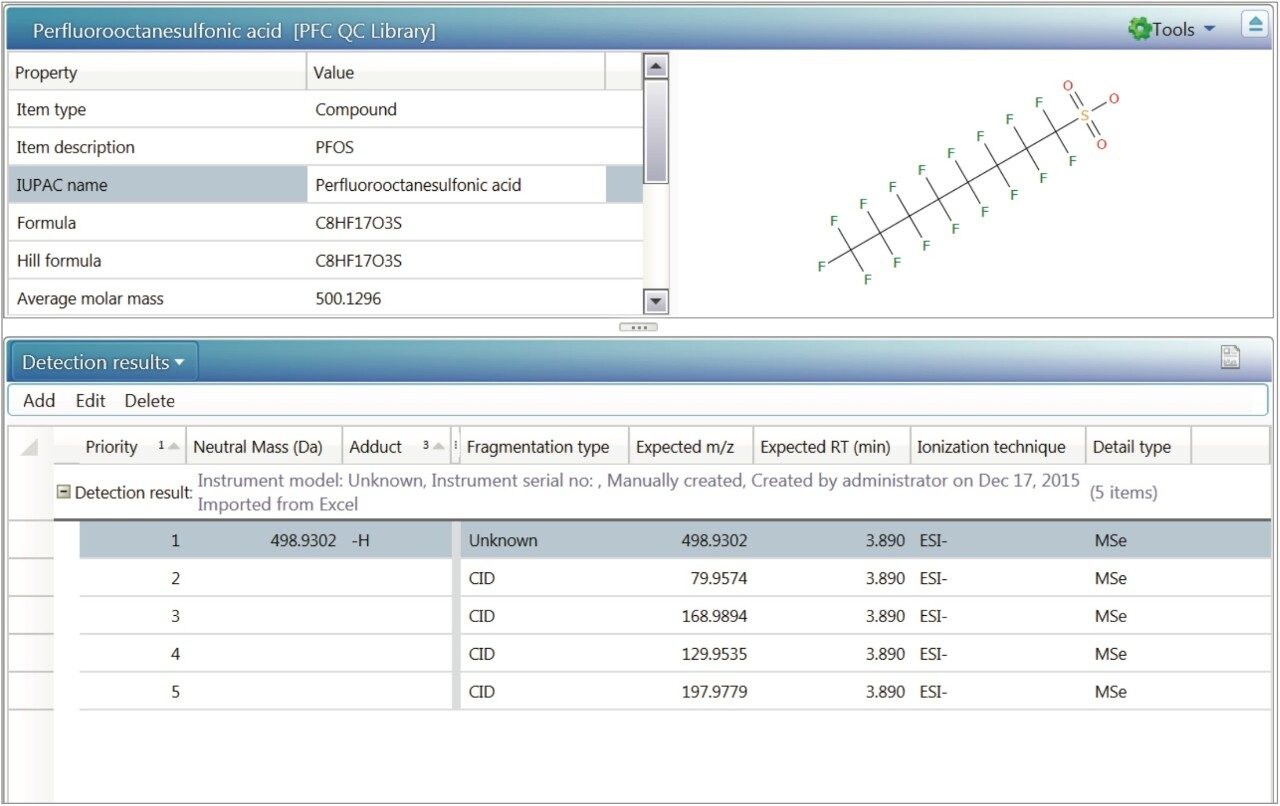
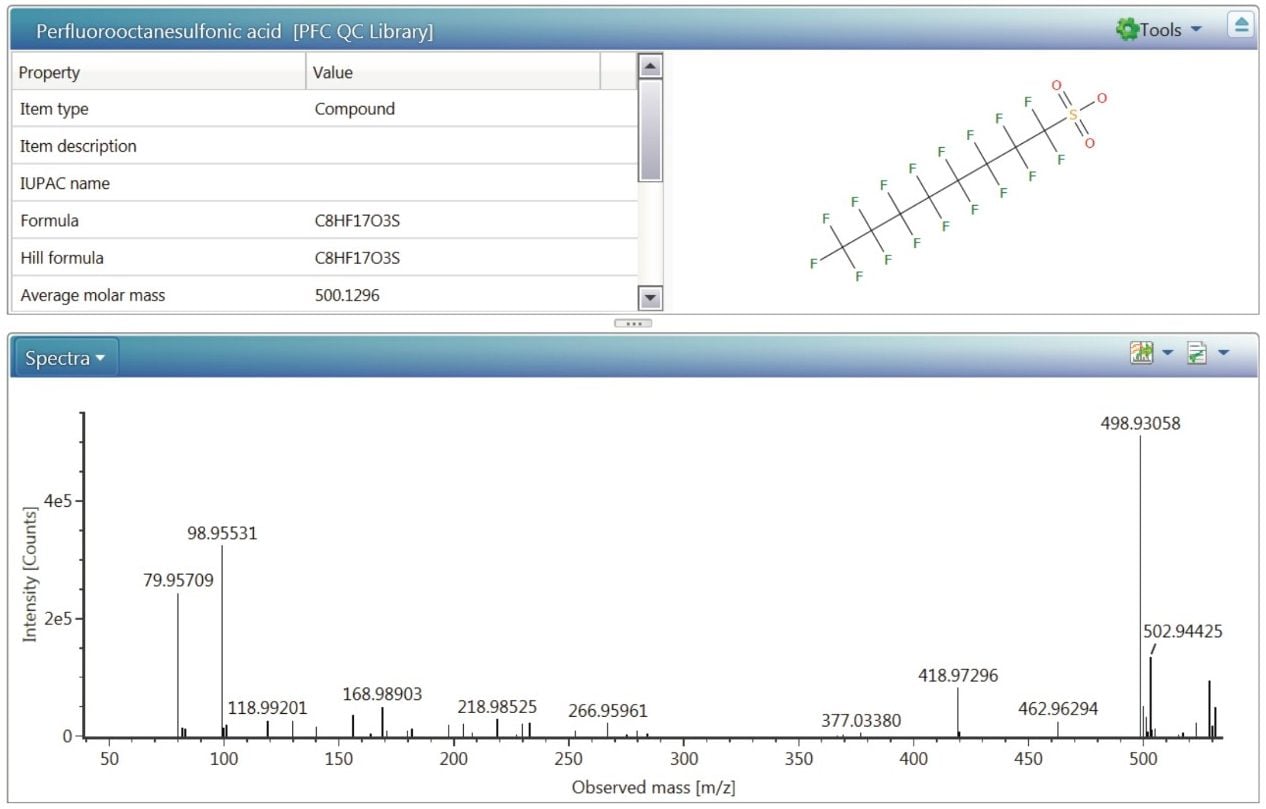
Componentization of the data7 allows users to interrogate a sample injection for target compounds and non-targeted (unknown) candidate masses of interest in the same dataset, without the need to reprocess the raw data. Information included in the target list comes from the scientific library functionality included in the UNIFI Scientific Information System. Libraries are user created and contain compound information such as structure, retention times and exact mass product ions, as well as capacity to store spectra and documentation about the compound. Figure 11 shows a library entry for PFOS, with detection results obtained from a standard injection.
In the case of emerging PFASs, or any other newly identified contaminants for which analytical standards may not be readily available, the use of compound structure can be used as a means to make tentative identifications. Because both low and high energy full spectral acquisition is achieved, UNIFI’s MassFragment tool will search not only for the precursor mass but also match proposed losses from the structure to masses observed in the high energy spectrum.
The UNIFI Scientific Libraries and target lists within an analysis method can be updated anytime, and the analysis re-interrogated to continually increase the numbers of compounds. This particular functionality is most useful in cases where the compounds are not yet found in online chemical databases, and users must generate their own structural information. Additionally, the Scientific Library functionality provides useful storage tools for experimental and compound information. Figure 12 shows a stored spectrum for PFOS, which can be used in future analyses to add confidence to a compound match.
Data processing using UNIFI’s Scientific Library functions greatly enhances the ability to rapidly identify expected as well as emerging compounds of interest. Historical data review is a powerful approach in particular for environmental and food analysis. It is made possible by full spectral acquisition and exact mass measurements of low and high collision energy data.
720005727, June 2016