This application note demonstrates how an integrated biopharmaceutical LC-MS system utilizing the UNIFI Scientific Information System addresses these challenges by integrating and automating data acquisition, data processing, and result reporting into a seamless workflow for in-depth biotherapeutic structural characterization.
For comparability studies performed with biosimilars, the integration of a fit-for-purpose UPLC/Tof-MS system with GxP-friendly data management, available with the UNIFI Scientific Information System, facilitates the development of a biotherapeutic product. This system solution enables complex biosimilar development to be carried out using routine analytical methodologies that are streamlined by efficient, workflow-based data management and reporting.
Biopharmaceutical companies are challenged to design efficient analytical strategies for detailed assessment of structural comparability between biosimilar and innovator products. Extensive characterization increases confidence that a biosimilar product is safe and will meet regulatory compliance requirements for abbreviated approval pathways. Here, we demonstrate how an integrated biopharmaceutical LC-MS system utilizing the UNIFI Scientific Information System addresses these challenges by integrating and automating data acquisition, data processing, and result reporting into a seamless workflow for in-depth biotherapeutic structural characterization.
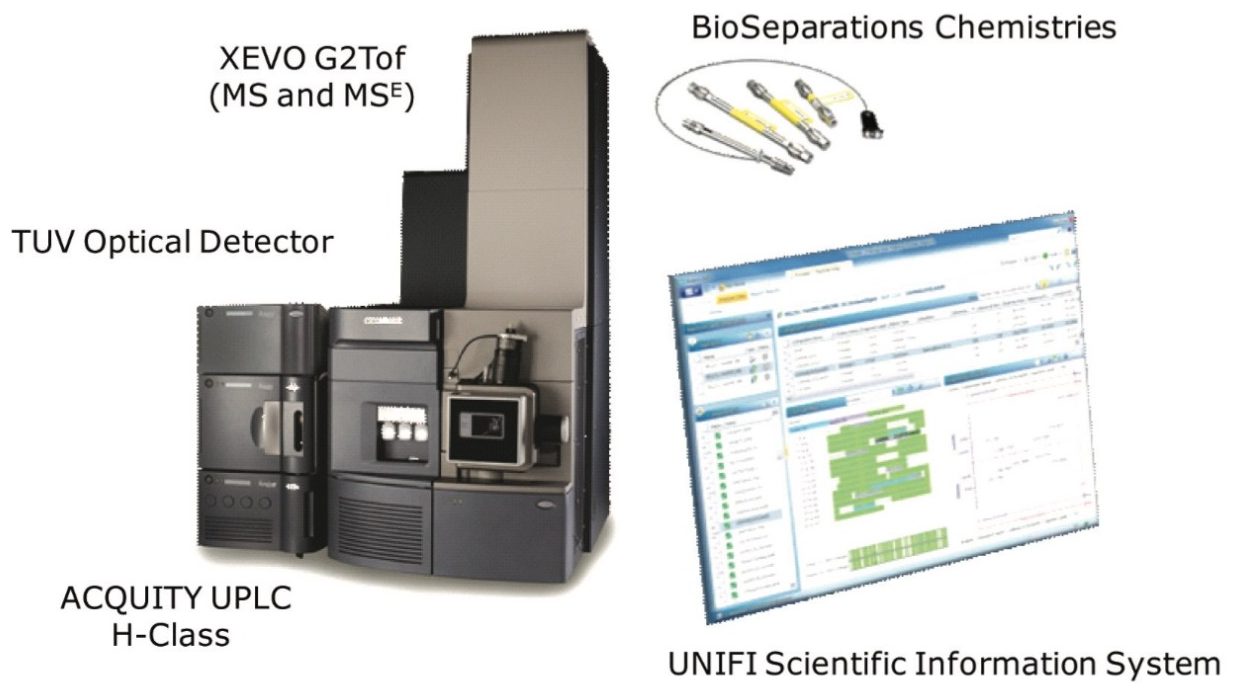
Comparability studies between an innovator, rituximab monoclonal antibody (mAb), and two biosimilar candidates were performed at the levels of intact protein, subunits (partially reduced antibody), and peptides using the Biopharmaceutical System Solution with UNIFI, shown in Figure 1. Differences in Critical Quality Attributes, such as primary structure (mutation), glycan fucosylation, and terminal amino acid heterogeneity were compared, quantified, and reported in a seamless workflow.
Intact mass analysis: Innovator and both of the biosimilar mAb samples were diluted to 0.5 mg/mL using 25 mM ammonium bicarbonate, pH 7.9 for injection and analysis.
Reduced mAb analysis: The samples were diluted to 1 mg/mL in a reduction buffer (25 mM NaCl, 25 mMTris, pH 7.5), and a concentrated DTT solution was added to the sample to obtain the final DTT concentration of 1.0 mM. The solution was then incubated at 37 °C for 20 min. The reduced samples were further diluted using a dilution buffer of 5% acetonitrile, 0.1% TFA to 0.2 mg/mL for LC-MS analysis.
Protein digestion: The samples were mixed with a denaturing buffer (8 M guanidine chloride, 1 M Tris, pH 7.5) to 1.0 mg/mL, reduced with 3 mM DTT, and alkylated with 7 mM iodoacetomide before buffer exchange over a NAP-5 column (GE Healthcare) to a digestion buffer of 100 mM Tris, pH 7.5. The samples were digested individually using either trypsin or chymotrypsin (S:E = 20:1) for 4 hrs. The digested samples were diluted with 3% acetonitrile, 0.1% TFA to 0.2 mg/mL for injection.
|
Biopharmaceutical System Solution with UNIFI: |
ACQUITY UPLC H-Class with Peptide Separation Technology (PST) and Protein Separation Technology (PrST) UPLC Chemistries Xevo G2 Tof, ACQUITY UPLC TUV Optical Detector UNIFI Scientific Information System |
|
Column: |
ACQUITY UPLC BEH300 C4, 2.1 x 50 mm |
|
Column temp.: |
80 °C |
|
Mobile phase A: |
water |
|
Mobile phase B: |
acetonitrile |
|
Mobile phase C: |
1% formic acid (aqueous) |
|
Detection: |
UV 280 nm |
|
Time (min) |
Flow (mL/min) |
%A |
%B |
%C |
%D |
Curve |
|---|---|---|---|---|---|---|
|
Initial |
0.3 |
85 |
5 |
10 |
0 |
Initial |
|
2.00 |
0.3 |
85 |
5 |
10 |
0 |
6 |
|
2.10 |
0.2 |
85 |
5 |
10 |
0 |
6 |
|
5.00 |
0.2 |
10 |
80 |
10 |
0 |
6 |
|
6.00 |
0.3 |
10 |
80 |
10 |
0 |
6 |
|
6.50 |
0.3 |
85 |
5 |
10 |
0 |
6 |
|
10.00 |
0.3 |
85 |
5 |
10 |
0 |
6 |
|
Capillary: |
2.5 kV |
|
Sampling cone: |
50 V |
|
Extraction cone: |
4 V |
|
Source temp.: |
150 °C |
|
Desolvation temp.: |
350 °C |
|
Cone gas flow: |
0 L/Hr |
|
Desolvation gas flow: |
800 L/Hr |
|
Column: |
ACQUITY UPLC BEH300 C4, 2.1 x 50 mm |
|
Column temp.: |
80 °C |
|
Mobile phase A: |
water |
|
Mobile phase B: |
acetonitrile |
|
Mobile phase C: |
1% formic acid (aqueous) |
|
Detection: |
UV 280 nm |
|
Time (min) |
Flow (mL/min) |
%A |
%B |
%C |
%D |
Curve |
|---|---|---|---|---|---|---|
|
Initial |
0.3 |
85 |
5 |
10 |
0 |
Initial |
|
2.00 |
0.3 |
85 |
5 |
10 |
0 |
6 |
|
2.10 |
0.2 |
85 |
5 |
10 |
0 |
6 |
|
3.00 |
0.2 |
65 |
25 |
10 |
0 |
6 |
|
13.00 |
0.2 |
60 |
30 |
10 |
0 |
6 |
|
13.10 |
0.3 |
10 |
80 |
10 |
0 |
6 |
|
15.00 |
0.3 |
10 |
80 |
10 |
0 |
6 |
|
15.50 |
0.3 |
85 |
5 |
10 |
0 |
6 |
|
25.00 |
0.3 |
85 |
5 |
10 |
0 |
6 |
|
Capillary: |
3.0 kV |
|
Sampling cone: |
30 V |
|
Extraction cone: |
4 V |
|
Source temp.: |
120 °C |
|
Desolvation temp.: |
350 °C |
|
Cone gas flow: |
0 L/Hr |
|
Desolvation gas flow: |
700 L/Hr |
|
Column: |
ACQUITY UPLC BEH300 C18, 2.1 x 150 mm |
|
Column temp.: |
65 °C |
|
Flow rate: |
0.2 mL/min |
|
Mobile phase A: |
water |
|
Mobile phase B: |
acetonitrile |
|
Mobile phase C: |
1% formic acid (aqueous) |
|
Detection: |
UV 214 nm |
|
Time (min) |
Flow (mL/min) |
%A |
%B |
%C |
%D |
Curve |
|---|---|---|---|---|---|---|
|
Initial |
0.2 |
89 |
1 |
10 |
0 |
Initial |
|
10.00 |
0.2 |
82 |
8 |
10 |
0 |
6 |
|
85.00 |
0.2 |
61 |
29 |
10 |
0 |
6 |
|
90.00 |
0.2 |
50 |
40 |
10 |
0 |
6 |
|
91.00 |
0.2 |
10 |
80 |
10 |
0 |
6 |
|
94.00 |
0.2 |
10 |
80 |
10 |
0 |
6 |
|
95.00 |
0.2 |
89 |
1 |
10 |
0 |
6 |
|
105.00 |
0.2 |
89 |
1 |
10 |
0 |
6 |
|
Capillary: |
3 kV |
|
Sampling cone: |
30 V |
|
Extraction cone: |
4 V |
|
Source temp.: |
100 °C |
|
Desolvation temp.: |
250 °C |
|
Cone gas flow: |
0 L/Hr |
|
Desolvation gas flow: |
500 L/Hr |
|
Column: |
ACQUITY UPLC BEH300 C18, 2.1 x 150 mm |
|
Column temp.: |
60 °C |
|
Mobile phase A: |
0.1% formic acid (aqueous) |
|
Mobile phase B: |
0.1% formic acid in acetonitrile |
|
Detection: |
UV 214 nm |
|
Time (min) |
Flow (mL/min) |
%A |
%B |
%C |
%D |
Curve |
|---|---|---|---|---|---|---|
|
Initial |
0.2 |
97 |
3 |
0 |
0 |
Initial |
|
1.00 |
0.2 |
97 |
3 |
0 |
0 |
6 |
|
91.00 |
0.2 |
57 |
43 |
0 |
0 |
6 |
|
91.10 |
0.2 |
25 |
75 |
0 |
0 |
6 |
|
94.10 |
0.2 |
25 |
75 |
0 |
0 |
6 |
|
95.00 |
0.2 |
97 |
3 |
0 |
0 |
6 |
|
98.00 |
0.2 |
97 |
3 |
0 |
0 |
6 |
|
Capillary: |
3 kV |
|
Sampling cone: |
30 V |
|
Extraction cone: |
4 V |
|
Source temp.: |
120 °C |
|
Desolvation temp.: |
350 °C |
|
Cone gas flow: |
0 L/Hr |
|
Desolvation gas flow: |
600 L/Hr |
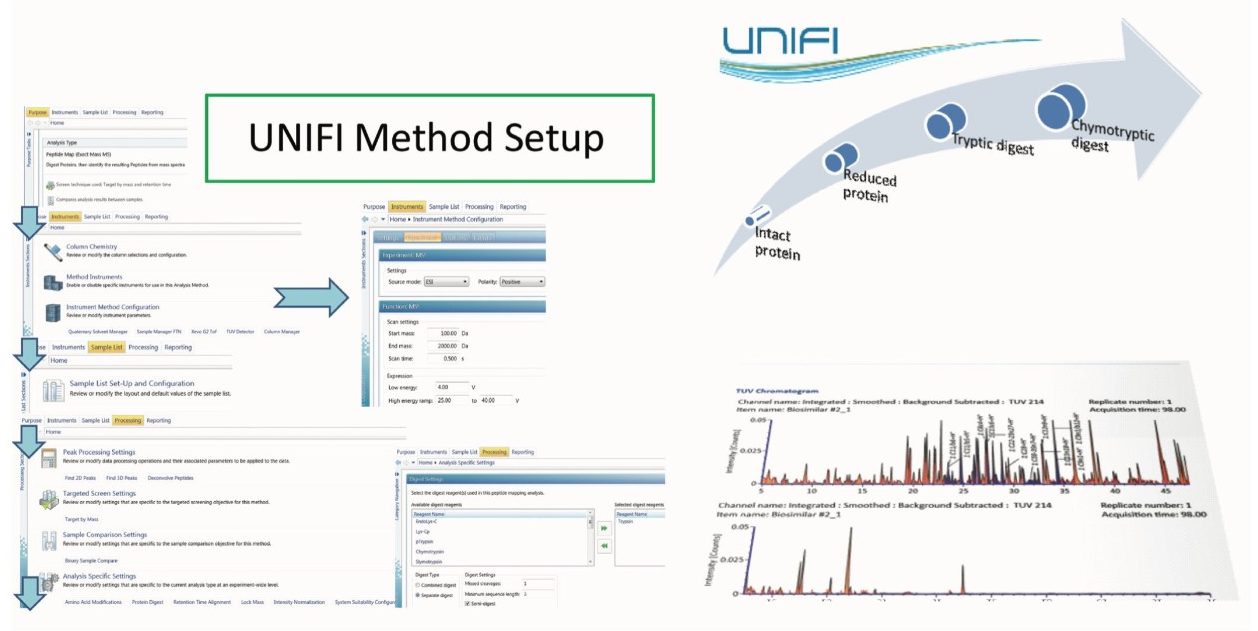
The therapeutic protein comparability workflow started with mAb sample analysis at the intact protein level, followed by the analysis of heavy and light chains after protein reduction, and finally addressed local post-translational modifications (PTMs) and mutations with LC-MSE peptide map methodology. Such comprehensive workflow is managed by UNIFI for a regulatory environment by integrating data acquisition, data processing, and reporting in a highly automated fashion. The analysis method is completely defined prior to acquisition with the instrument settings, data processing parameters, and a reference to a reporting template included. Each analysis type focuses on a particular application need, such as intact protein analysis or peptide mapping experiment, facilitating the design of a method workflow, as shown in Figure 2. The report templates are composed of the objects that can be entirely configured by the user. The standard report templates include total ion chromatogram (TIC), mass spectra for all or selected ions in a form of either raw, deconvoluted, or centroid data format, and a tabulated summary of the interpreted LC-MS(MSE) data.
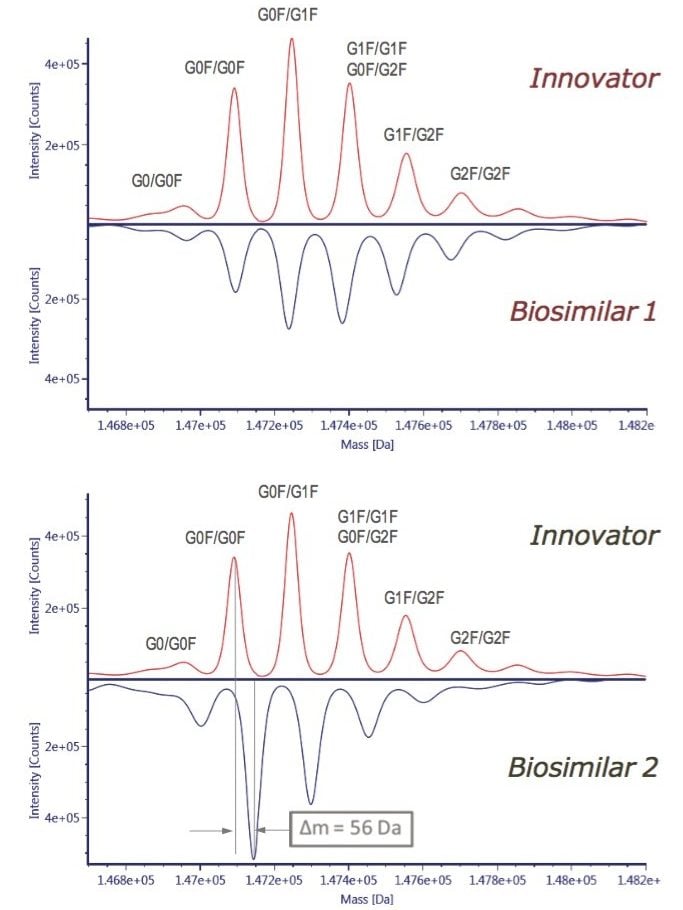
For a quick assessment of the possible differences among the innovator, rituximab, and two biosimilar samples (Biosimilar 1 and Biosimilar 2), the intact protein mass analysis was performed. UNIFI has a built-in MaxEnt1 deconvolution capability for protein MW calculation and comparison. Figure 3 shows the distribution of glycoforms on the deconvoluted mAb spectra presented as the mirror plots. A systematic mass shift of 56 Da was observed in Biosimilar 2 glycoforms with respect to the innovator mAb; whereas, the Biosimilar 1 glycosylation profile displayed inconsistent mass difference (except G0F/G0F glycoform). The intact protein analysis data can also be viewed as raw, centroid spectra, or as a component summary, and can be used for the first-round evaluation of mAb sample heterogeneity.
A closer look at the reduced form of rituximab allowed users to confine the structure heterogeneity to the individual heavy or light protein chains. Partially reduced mAb analysis measured and compared PTM and glycosylation profile among the innovator and both biosimilar mAbs, as seen in Figure 4. Consistent with 56 Da mass shift observed from intact protein data, our data suggest that 28 Da mass difference, possibly an amino acid sequence variation, belongs to the heavy chain of Biosimilar 2. N-terminal pyroglutamination Q (PyrQ) levels were measured and reported for heavy and light chains. C-terminal Lys variants on the heavy chain as well as glycoform variants were automatically assigned in the UNIFI Review panel and plotted across all the samples of the innovator and biosimilars.
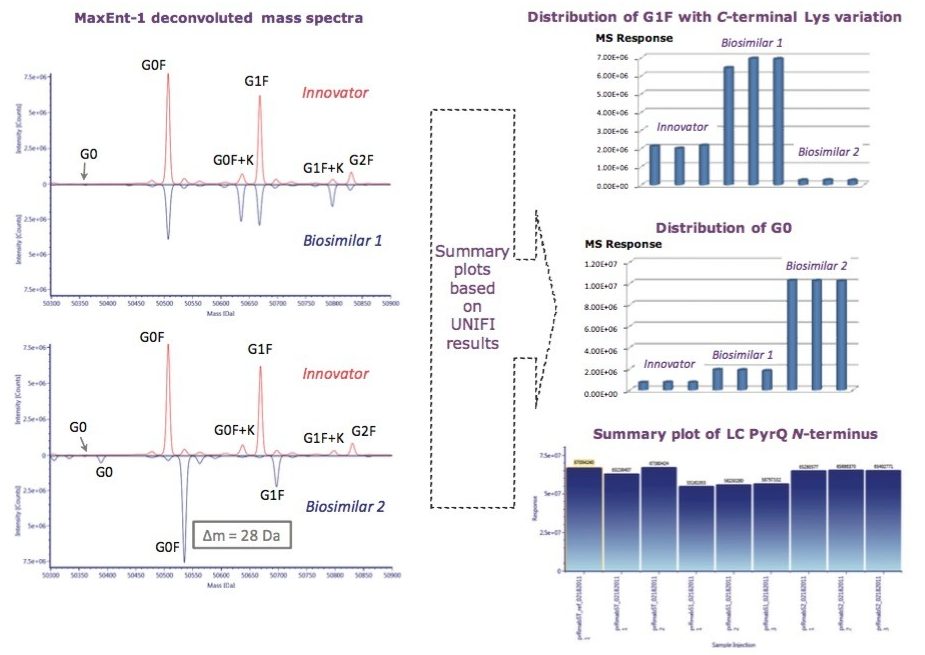
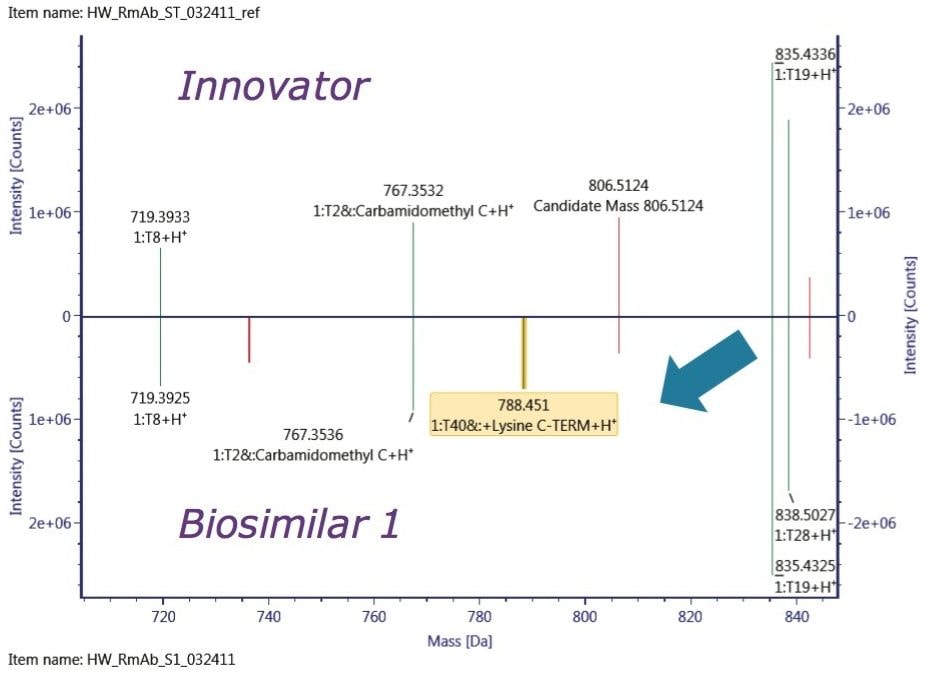
In comparison with the innovator, the obvious difference displayed in Biosimilar 1 in the deconvoluted HC spectrum is the higher degree of C-terminal Lys variation, which contributes to the inconsistent mass shift observed in the intact mass analysis. One of the remarkable differences between the two biosimilars was the relative abundance of G0 glycoform, which is known to correlate with antibody-dependent cellular cytotoxicity,1 and is believed to affect drug safety and efficacy.
The summary plot tool allows users to select any observable data, such as response, mass error, retention time, etc., and trend it across all the injections, which is one of the UNIFI assets of the automatic and efficient data reviewing.
To localize the difference among the three mAb samples, peptide mapping data were collected. A mirror plot of the tryptic digest demonstrated C-terminal Lys variant exists only in the Biosimilar 1 peptide map, shown in Figure 5, which was consistent with the glycosylation profiling data at the reduced protein level.
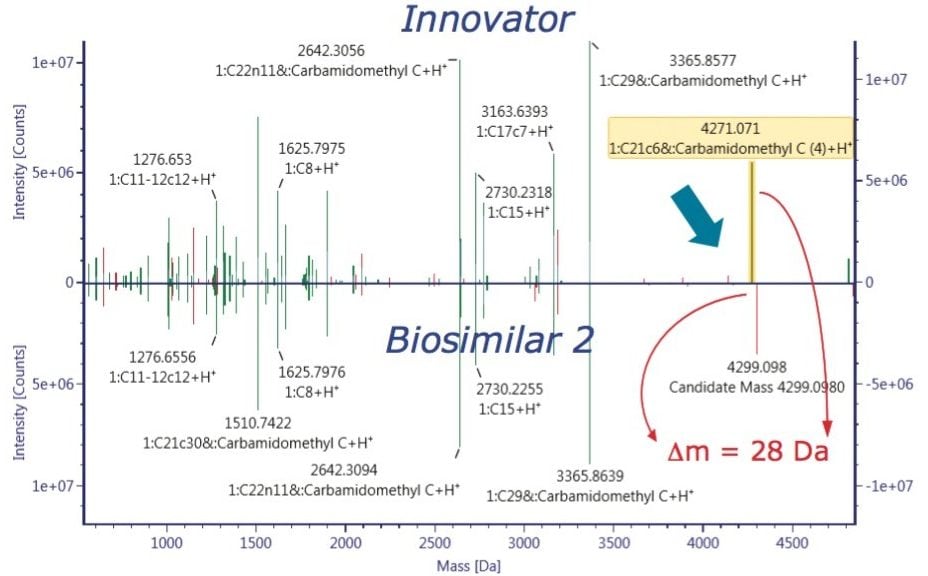
The ultimate inquiry was localizing the amino acid mutation contributing to 28 Da mass shift of the Biosimilar 2. Based on published information,2 an additional targeted sequence with Lys218 → Arg218 mutation was submitted to the method search. Compare mode view of the tryptic digest chromatogram, or component summary did not show a significant difference between the innovator and Biosimilar 2 mAb; therefore, no conclusion could be drawn about the primary sequence difference. The answer came with use of an alternative, non-specific enzyme, chymotrypsin. The chymotryptic map clearly showed the mass shift in a component view, as seen in Figure 6, and the peptide with a mutation site was automatically highlighted in the chromatogram, peptide map, and the component summary in the Review panel. Filtering the results in the Review by “showing unknown unique components” makes it easy to display the differences between the innovator (“reference”) and the biosimilar samples (“unknown”).
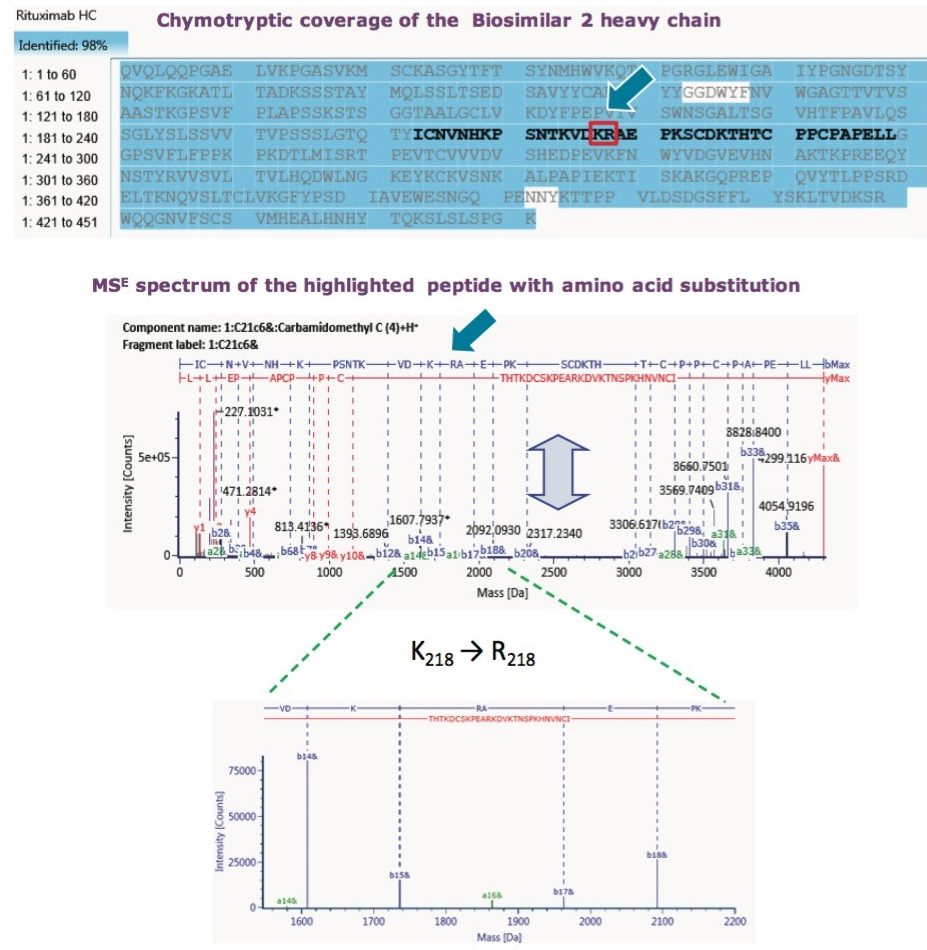
The reason that the tryptic map failed to pinpoint the amino acid substitution is that proteolytic cleavage occurs at Lys217, Lys218 or Arg218. So, the very amino acid of question gets cleaved as a single amino acid entity. Chymotryptic digest, on the other hand, captures the mutation within a single peptide. Finally, Lys218 → Arg218 substitution was confirmed with MSE data, as seen in Figure 7, which displayed a16-ion fragment characteristic of Arg. UNIFI peptide map workflow proved the capability to confirm sequence mutation or other suspected PTMs.
UPLC/TOF-MS analysis at intact mAb, reduced mAb, and peptide map levels enabled the detection of primary structural differences, and quantitative assessments of these variations. An integrated biopharmaceutical LC-MS system utilizing the UNIFI Scientific Information System with automated data acquisition, processing, and reporting for multiple analytical workflows enabled the efficient assessment of critical product attributes with minimal manual intervention.
The K → R mutation found in the Biosimilar 2 (of rituximab) study is not readily detectable under tryptic digest analysis. It demonstrates the need to routinely employ alternative digestion enzymes for product characterization. The integrated workflow of protein characterization at different levels, combined with intelligent methods and tools of UNIFI, will improve productivity and cut the cost of biosilimar drug development.
720004445, September 2012