
This is an Application Brief and does not contain a detailed Experimental section.
This application brief demonstrates to successfully carry out broad scope pesticide screening in food commodities using ToF/MS; with increased confidence in the reported results arising from the reproducibility of mass accuracy across detected peaks, even at low concentrations.
Xevo G2 QTof acquired robust and reliable MSE data, with exact mass RMS variation of less than 2 ppm for both precursor and product ions at a concentration less than half that of the MRL.
Pesticides are used to generate improved crop yields as the demands on global food production increase. The quality of the food we eat is very carefully regulated worldwide, and a key component of food safety legislation is ensuring that pesticide residues do not remain on crops sold to consumers, as this could pose a health risk.
Recently, European legislators updated the guidance for pesticide residue analysis in food and feed,1 which now includes the recognition that accurate mass data may be used as part of the evidence for the presence or absence of analytes of interest. Consequently, data acquired on a time-of-flight (TOF) instrument can be used when reporting findings from pesticide screening analyses of food crops.
Typically, a TOF-screening approach might be used to help reduce the number of suspected positive compounds, prior to the sample being analyzed using a targeted MRM method on a tandem quadrupole instrument. However, reducing the number of false positives is essential to ensure that this approach is both robust and reliable. TOF screening also provides the ability to re-interrogate historical data for compounds not previously targeted, which offers added benefit compared with targeted MRM data.
Xevo G2 QTof MS coupled with ACQUITY UPLC, along with the Waters TOF Screening Pesticide Database, and POSI±IVE Software data processing, were used to rapidly screen extracted green bean samples. An incurred residue of the fungicide thiabendazole was detected, as illustrated in Figure 1.
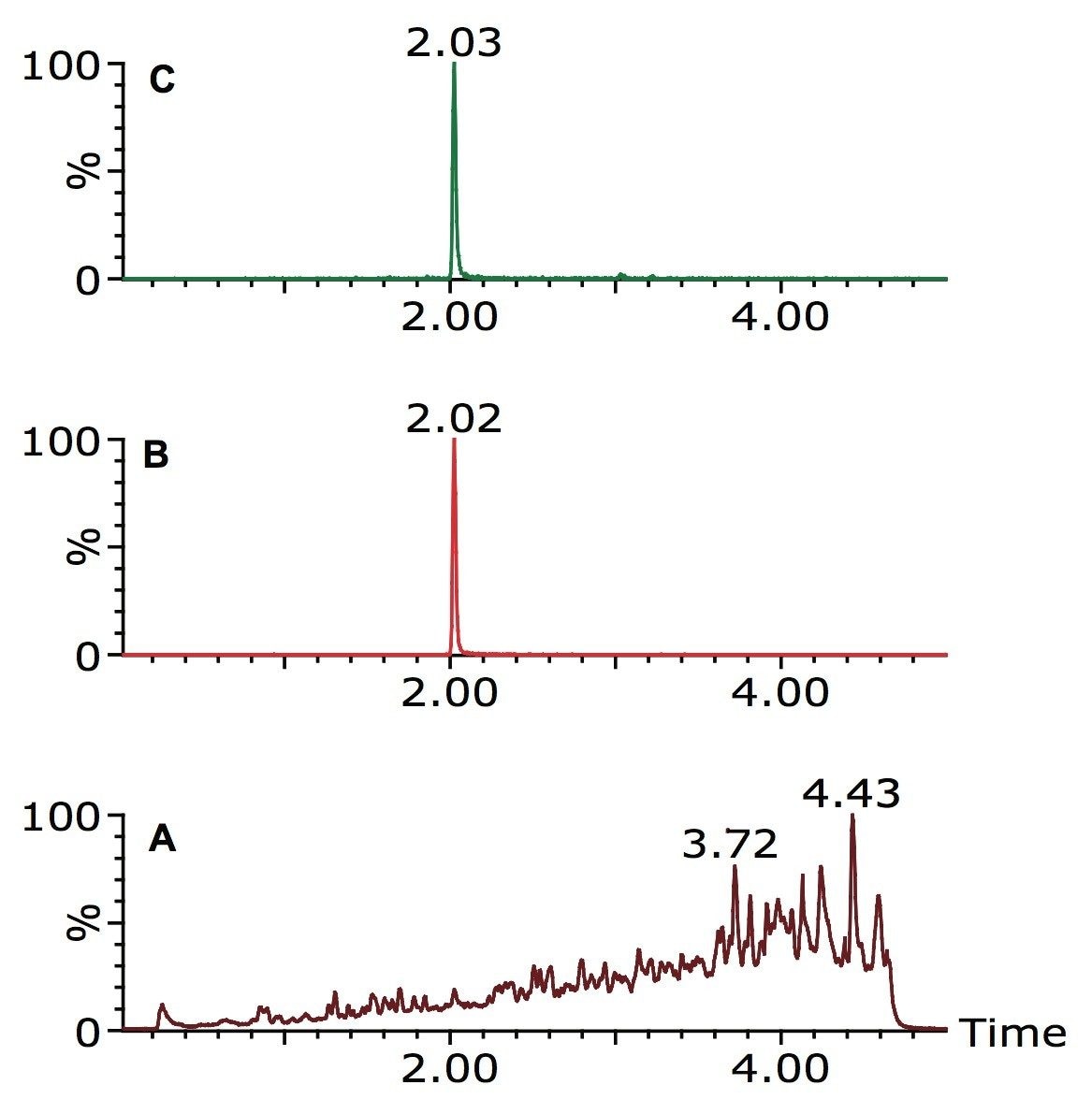
The MSE functionality of the Xevo G2 QTof Mass Spectrometer enables the acquisition of both low energy precursor ion and high energy fragment ion mass spectral data in a single run. A generic screening UPLC gradient was used, with a total runtime of five minutes. The mobile phases used were 10 mM ammonium acetate solution in water and 10 mM ammonium acetate solution in methanol.
The extracted ion chromatograms in Figure 1 have 12 points across the low energy precursor ion peak, and 12 points across the high energy fragment ion peak, giving a total of 24 data points for thiabendazole. In addition, the Xevo G2 QTof Mass Spectrometer provides extremely accurate exact mass data for not only the precursor ions, but also the fragment ions.
Table 1 illustrates the exact mass data for thiabendazole in extracted green beans. The calculated RMS values for exact mass variation for both precursor and fragment ions were well below 2.0 ppm, even for these low mass ions.
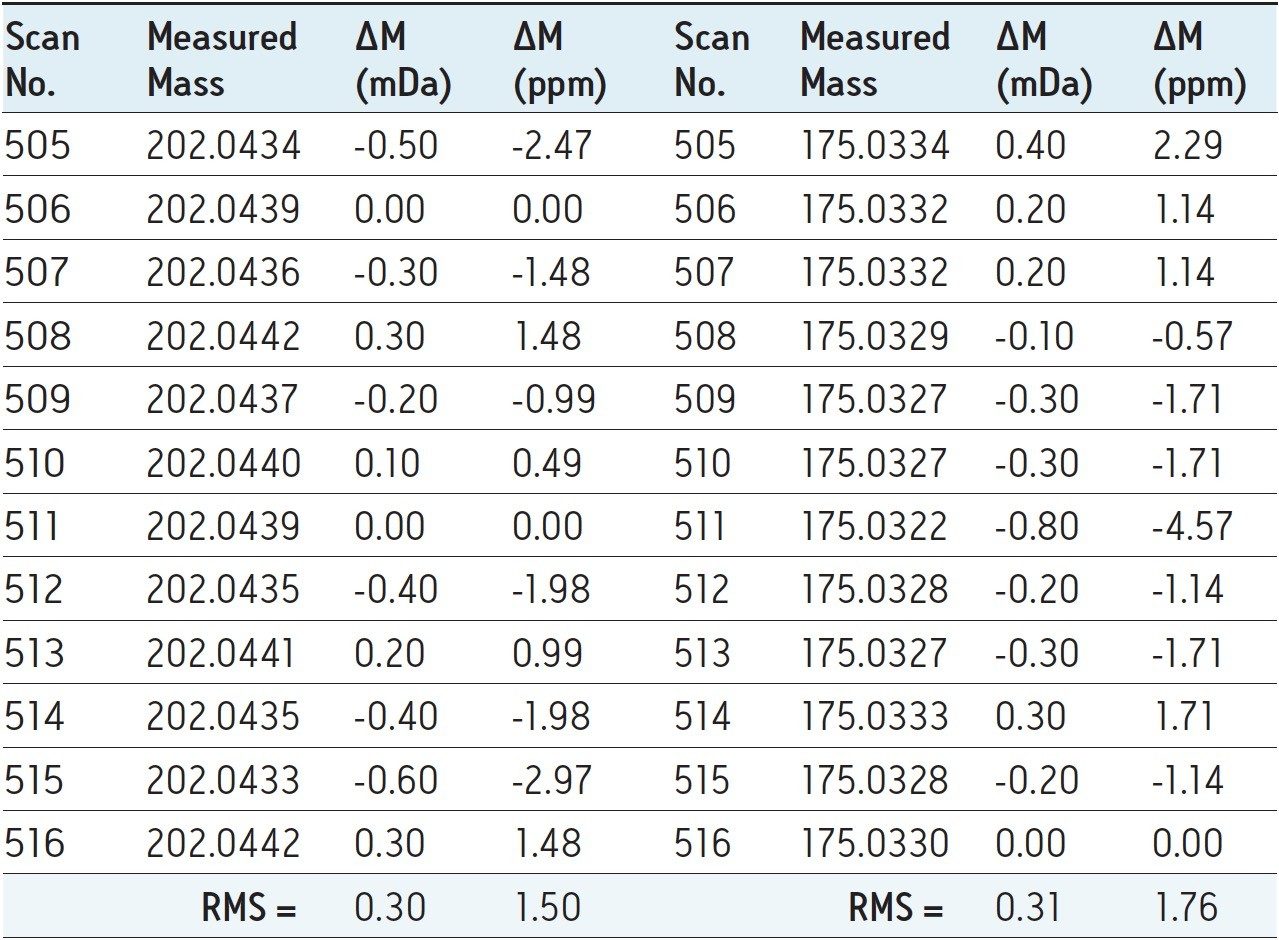
The EU Maximum Residue Limit2 (MRL) for thiabendazole in green beans is 0.05 mg/kg (50 μg/kg), and the data presented here is at a concentration less than half that of the MRL, demonstrating robust and reliable exact mass data at a low analyte concentration.
The Xevo G2 QTof together with ACQUITY UPLC was successfully used to screen extracted green beans for pesticide residues. An incurred residue of thiabendazole was discovered at a concentration below that of the MRL but well above the Limit of Detection (LOD) of the instrument.
Use of the MSE functionality of the Xevo G2 QTof enabled the acquisition of exact mass data for both precursor and fragment ions in one screening run, with a high level of reproducibility. This provides extra confidence in the identification of incurred residues and reduces false positives.
A reduction in false positives streamlines the workflow when transferring the samples to a tandem quadrupole instrument for confirmatory analysis, and provides valuable time-saving and efficiency benefits to the cost conscious analytical laboratory.
Waters offers a complete solution for broad scope TOF screening for food commodities, comprised of DisQuE sample extraction with ACQUITY UPLC separation and Xevo G2 QTof detection, followed by data processing with POSI±IVE Software.
720003421, April 2010