In this work, the SPE screening protocol and method development plate were successfully used to develop extraction methods for none out of 12 peptides tested on a first pass.
Biomolecules such as proteins and peptides represent a growing number of pharmaceutical drug entities. Drugs in this class – biop-harmaceuticals – are often more specialized than small molecules since they typically replace or augment an endogenous molecule known to change a disease or physiological state.1 Furthermore, since they are analogs to, synthetic versions of, or are endogenous compounds, they tend to be well-tolerated by the body. There are currently more than 40 peptide-based drugs on the market and more than 400 in late-stage clinical trials.2
Development of bioanalytical methods for the detection of small molecule pharmaceuticals in humans and animals is a challeng-ing and time-consuming process. Regulatory guidelines require methods to be acceptable in terms of linearity, sensitivity, accuracy and precision, selectivity, stability, and carryover. Bioanalytical methods now need to be developed for biopharmaceuticals as well – and many of the same challenges faced in bioanalysis of small molecules also apply to these larger biopharmaceuticals.
Achieving selectivity for analytes in complex biological matrices is the key component of all bioanalytical methods. Selective sample preparation continues to play a critical role in minimizing matrix effects and reducing variability associated with incurred samples. In the extraction solution we describe here, mixed-mode solid phase extraction (SPE) is employed for this purpose, since it relies on both reversed-phase and ion exchange mechanisms. Mixed-mode SPE selectively separates the desired analytes from matrix components and has been shown to reduce matrix effects.3
Developing bioanalytical methods for peptides is further compli-cated by several factors. Peptides are zwitterionic, making their retention behavior on SPE sorbents difficult to predict and therefore increasing method development times. Because of their size and charge state distribution, sensitivity by mass spectrometry may be lower for biomolecules than typical small molecules. This often necessitates concentration during sample preparation.
The Waters Peptide Method Development Kit was designed to facilitate the development of selective extraction and liquid chromatographic methods for peptides in biological matrices. The Peptide Separation Technology Method Development Kit is available for ACQUITY UPLC (p/n: 176001835) or HPLC (p/n: 176001836) analysis. The kit contains a chromatographic column, an Oasis µElution Peptide Method Development extraction plate, a simple and straightforward SPE screening protocol, and an LC screening protocol.
In this application note, the kit was used to develop extraction and chromatographic methods for the 12 peptides described in Table 1.


Spiked human plasma samples were pre-treated by diluting 1:1 (v:v) with 4% H3PO4. Samples were then extracted using the Oasis µElution Peptide Method Development extraction plate according to the basic screening protocol included in the kit. The ACQUITY UPLC System was used to increase the sensitivity of the separation and resolution between analytes and similar endogenous compounds required for a selective assay. Samples were analyzed with the ACQUITY UPLC Peptide Column included in the kit. Peptide columns are quality-control tested with peptide standards to ensure consistent performance.
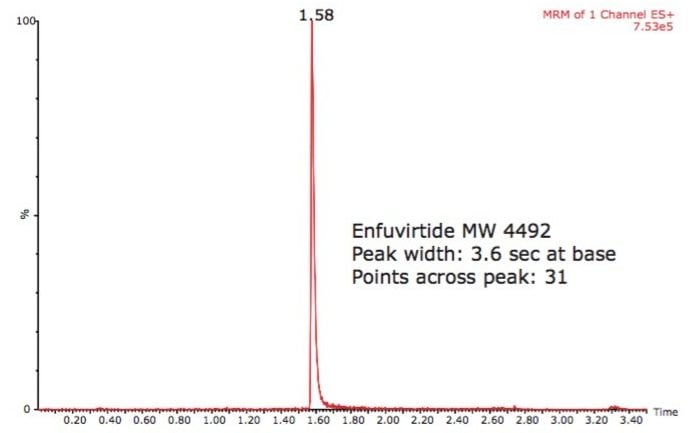
The Xevo TQ MS tandem quadrupole mass spectrometer was used to detect the peptides in positive ion electrospray. Parent masses were detected for the peptides in their 2+, 3+, or 4+ charge states and fragments were either singly or multiply charged. Since UPLC peak widths were on the order of 2 to 4 seconds wide at base for all peptides, it was important to have a mass spectrometer capable of not only providing enough points across the peaks for accurate and reproducible quantitation, but also one that could maintain this performance under constantly changing collision cell conditions encountered when monitoring multiple peptides at once.
Another key MS requirement for peptide bioanalysis is mass range. The mass range of both quadrupoles should be at least 1500 amu, preferably to 2000 amu, to accommodate higher m/z charge state precursors and fragments, which may provide the highest sensitivity for larger peptides. The Xevo TQ MS was chosen for its superior performance in these areas.
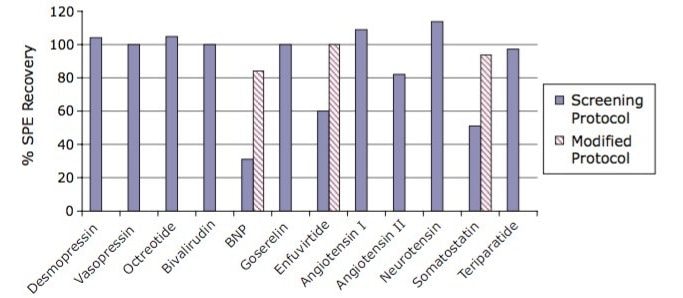
The screening protocol was developed to streamline the choice of SPE sorbent and to provide initial SPE recovery and matrix effect data for the analytes of interest. For many peptides, analyte recov-ery and matrix effects resulting from the basic screening protocol were acceptable. Matrix effects, where measured, were less than 11%. Absolute matrix effects of less than 15% were considered acceptable for this study. The negligible level of matrix effects observed indicates the specificity of the methodologies.
Recovery for the peptides from human plasma was assessed by comparing peak areas from pre-spiked extracted samples to blank plasma extracts spiked post-SPE.
The screening protocol yielded greater than 82% recovery for nine out of 12 peptides extracted from human plasma. Minor modifications made to the protocol improved recovery for the three other peptides, to more than 83%. Figure 2 summarizes analyte recovery for the diverse set of 12 peptide therapeutics extracted using the screening protocol. The extraction was carried out in a 96-well Oasis µElution Plate format to facilitate direct injection of the extracted sample and to obtain the analyte concentration necessary to meet required LOD/LLOQs.
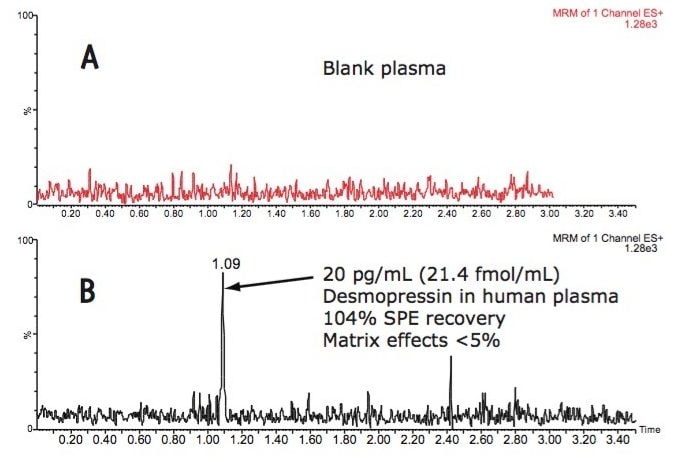
A concentration factor of up to 15X can be achieved without evaporation using the µElution format. It is particularly desirable to eliminate the evaporation step when analyzing peptides as this minimizes any evaporative losses or adsorption to the walls of collection plates. A representative chromatogram for desmopressin extracted from human plasma is shown in Figure 3.
Chromatography was performed using the UPLC screening method provided in the kit. The chromatographic method has a cycle time of 3.5 minutes, enabling hundreds of samples to be analyzed per day. Sample chromatograms for two of the peptides are shown in Figures 4 and 5.
The behavior of peptide therapeutics under various sample prepara-tion conditions may be difficult to predict. This has the potential to make development of a selective sample prep method compli-cated and time consuming. Using the Oasis Peptide µElution Method Development Plate and protocol greatly simplifies extraction method development and rapidly identifies an appropriate SPE sorbent and starting protocol.
In this work, the SPE screening protocol and method development plate were successfully used to develop extraction methods for nine out of 12 peptides tested on a first pass. Minor modifications to the screening protocol resulted in suitable extraction methods for the three other peptides.
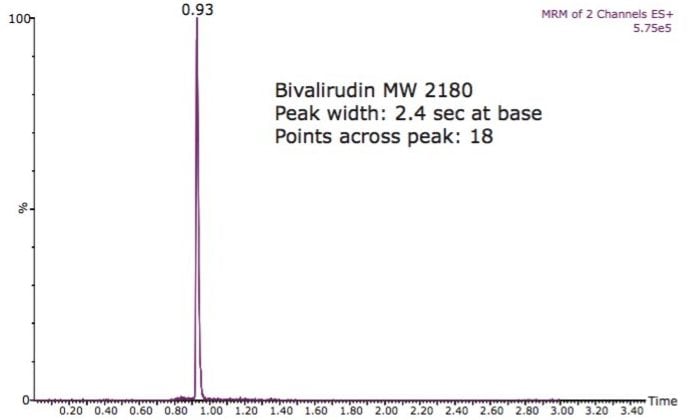
Peaks widths on the order of 2 to 4 seconds wide at base and more than 15 points across the peak were obtained for all peptides tested using the ACQUITY UPLC Column provided in the kit.
Overall, we have shown that bioanalysis studies for peptide therapeutics are amenable to a platform-based approach to method development. Such standardized approaches for determining optimal SPE recovery and concentration and MRM-based LC-MS analysis enables laboratories to reduce development timelines and shorten time to market for peptide drugs.
720003253, October 2009