
This application note describes the use of the ACQUITY UPLC System, the OST Column, and the Q-Tof Premier Mass Spectrometer for the study of oligonucleotides. This UPLC-MS methodology demonstrates the ability to use Waters technology to quantitate oligonucleotides while providing significant structural information, resulting in both improved quality as well as productivity for biopharmaceutical laboratories, making UPLC/MS an enabling technology for analysis of DNA/RNA based therapeutics.
Synthetic oligonucleotides are used extensively in the field of molecular biology, clinical diagnosis, and the development of new therapeutic agents. Quantitative and qualitative methods are required for the analysis of these oligonucleotides.
With a growing number of antisense- and RNAi-based drugs in development and clinical trials, a reliable and sensitive liquid chromatography method with mass spectrometry detection (LC-MS) is highly desirable.
The inherently unique characteristics of therapeutic oligonucleotides combined with the multiple-step manufacturing process make analysis of these oligonucleotides challenging. Post-purification analysis is a difficult and time-consuming process, typically requiring multiple orthogonal methods (CGE and SAX HPLC), adding significant costs and burden to an analytical laboratory. Furthermore, CGE and SAX HPLC are unable to resolve and quantitate many of the process-related impurities and degradent products that may exist after primary purification. Additionally, neither technique can provide significant structural data about the oligonucleotide, requiring the use of additional techniques.
The Waters ACQUITY UltraPerformance (UPLC) System combines with Oligonucleotide Separation Technology (OST) Columns, packed with 1.7 μm sorbent, to provide superior analytical performance for oligonucleotide separations compared to HPLC and fast LC separations.
This application note describes the use of the ACQUITY UPLC System, the OST Column, and the Q-Tof Premier Mass Spectrometer for the study of oligonucleotides. This methodology demonstrates outstanding separation efficiency and sensitivity together with high mass accuracy, resulting in an improved quality and high throughput analysis.
|
LC system: |
Waters ACQUITY UPLC System |
|
Column: |
ACQUITY UPLC OST C18 2.1 x 50 mm, 1.7 μm |
|
Column temp.: |
60 °C |
|
Flow rate: |
0.2 mL/min |
|
Mobile phase A: |
15 mM TEA. 400 mM HFIP |
|
Mobile phase B: |
50% A, 50% methanol |
|
Gradient: |
38 to 48% B in 10 min |
|
Detection: |
ACQUITY UPLC PDA, 260nm |
|
MS System: |
Waters Q-Tof Premier Mass Spectrometer |
|
Capillary: |
2500 V |
|
Sample cone: |
35 V |
|
Extraction cone: |
3 V |
|
Ion guide: |
2.5 V |
|
Desolvation temp.: |
200 °C |
|
Source temp.: |
120 °C |
|
Cone gas flow: |
50 L/hr |
|
Desolvation gas flow: |
600 L/hr |
|
Lock mass: |
10 mg/mL, CsI, 5 μL/min |
|
Scan time: |
1 sec |
|
Frequency: |
30 sec |
A MassPREP OST standard (PN 186004135) consisting of 15, 20, 25, 30, and 35 nt (nucleotides) long oligodeoxythymidines was used as a sample to demonstrate the performance of ACQUITY UPLC System, OST Columns, and the Q-Tof Premier Mass Spectrometer. The approximate quantity of each oligomer in the vial is listed in Table 1. The oligomers were reconstituted in 500 μL of 0.1 M triethylamine acetate (TEAA) before LC injection.
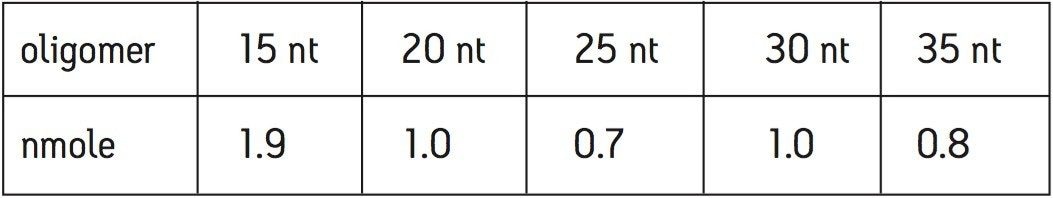
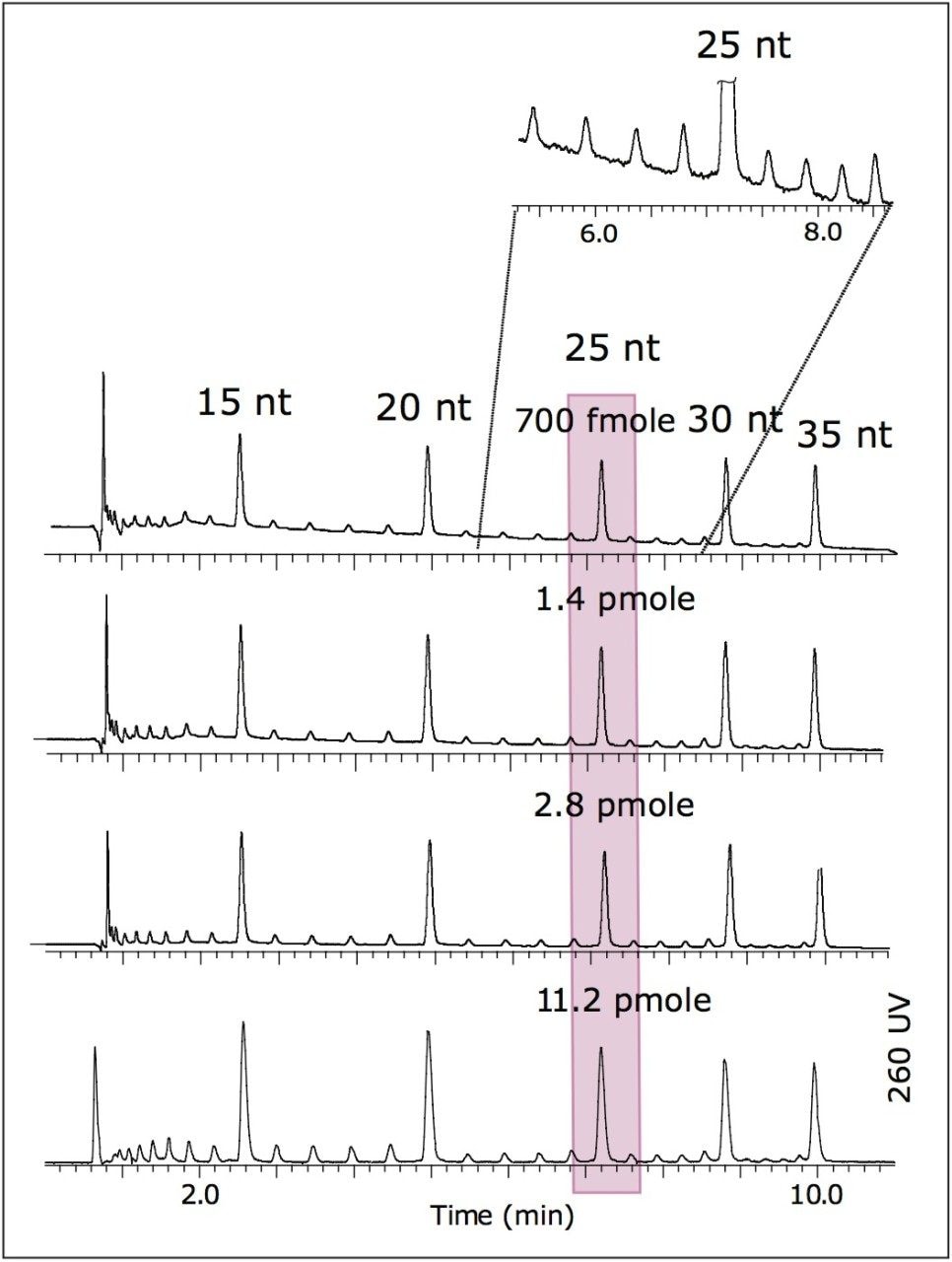
UPLC separation of oligonucleotides was performed with MS-compatible mobile phases comprised of aqueous solution of 15 mM triethylamine (TEA) and 400 mM hexafluoroisopropanol (HFIP), pH 7.9, and methanol. The resulting chromatogram shows an efficient separation of 15, 20, 25, 30, and 35 nt oligonucleotides from the by-products of synthesis, customarily termed failed sequences (Figure 1).
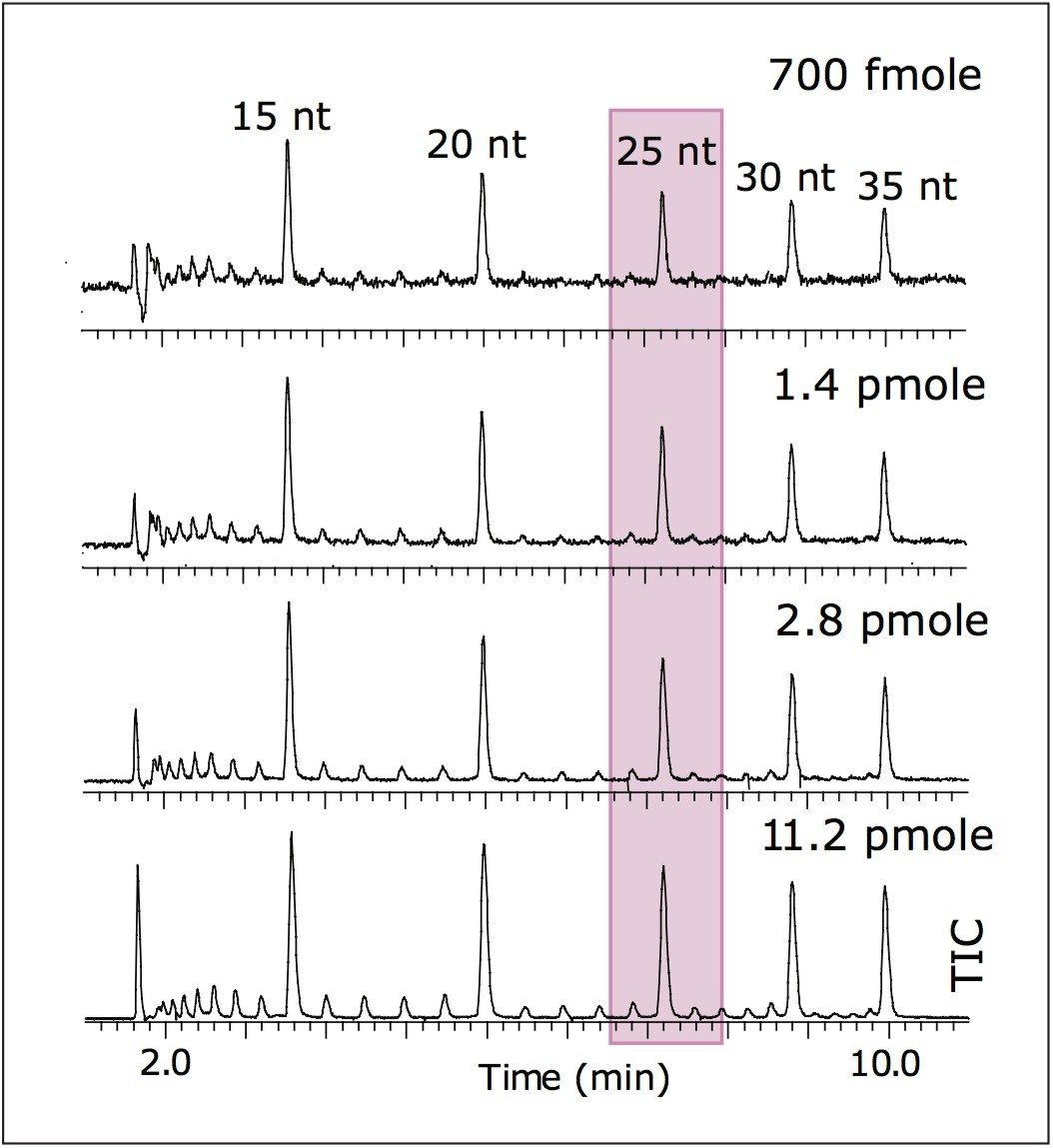
Both UV and MS detection was used in series. The ACQUITY UPLC PDA detector was connected to the Q-Tof Premier using 75 μm x 70 cm silica capillary tubing. The MS scan time was 0.45 sec to collect at least 20 data points across the chromatographic peak.
The UV limit of quantitation (LOQ) given chromatographic system (S/N=10) at UV 260 nm was estimated from Figure 1. The LOQ for 26 nt was ~70 fmoles.
Mass spectrometry data were acquired with the Q-Tof Premier operating in negative ion mode. Results are shown in Figure 2. The LOQ estimate for MS was ~700 fmoles (25 nt). This is more than sufficient for MS analysis of minor peaks corresponding to the failed sequences of oligonucleotides.
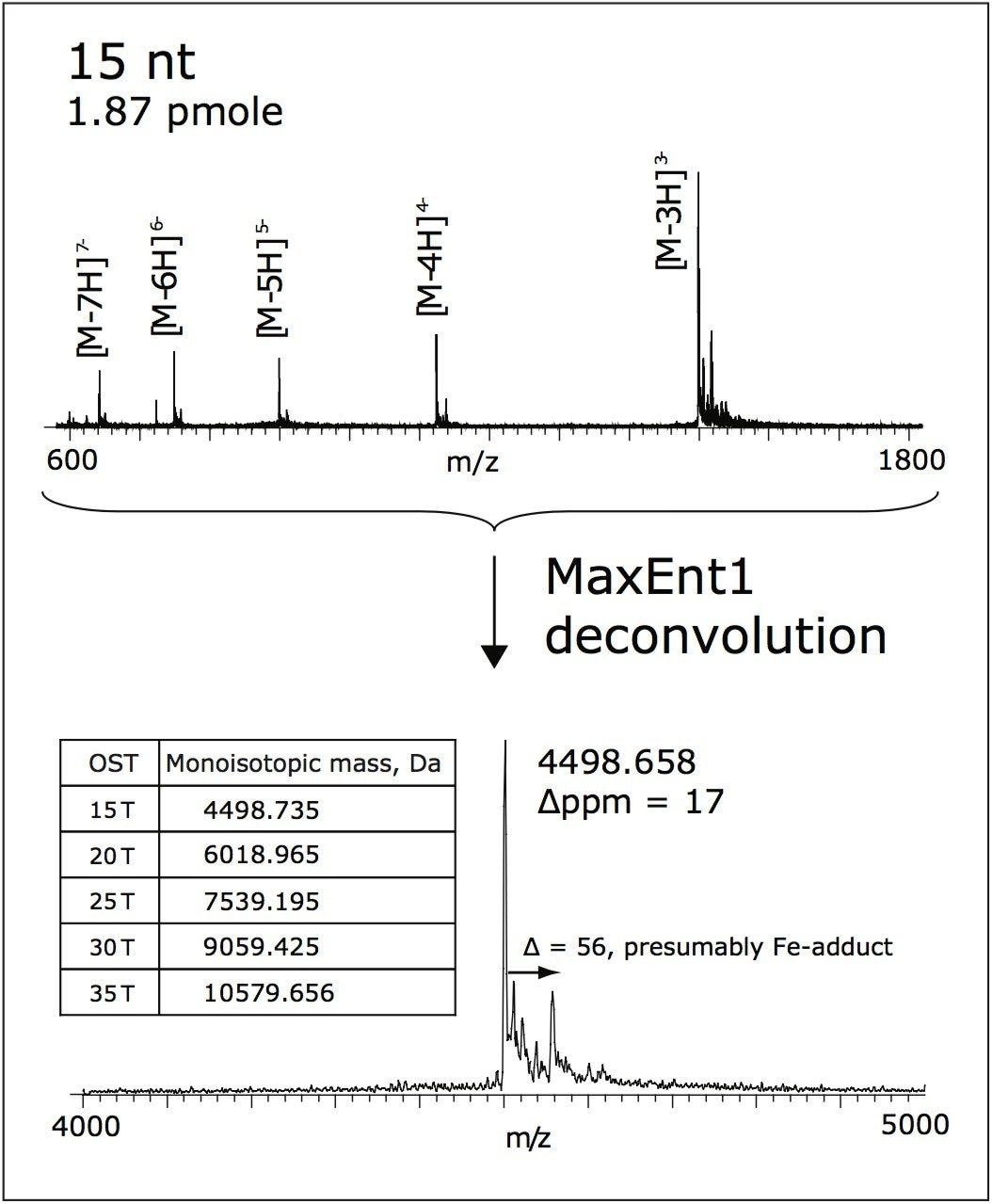
Average mass accuracy of 17 ppm was obtained for the OST Column (15 nt), internally calibrated by “lock-mass” method, as shown in Figure 3. CsI (10 mg/mL in isopropanol/water, 1:1) was used as the lockmass reference.
The Q-Tof Premier Mass Spectrometer has been applied for sensitive analysis of synthetic oligonucleotides following high resolution UPLC separation. The ACQUITY UPLC OST Column, gradient conditions, and MS parameters were designed to perform high-throughput and reproducible separations, with sensitive and accurate mass detection. Sub-picomole LOQ’s were achieved using the proposed method.
This UPLC-MS methodology demonstrates the ability to use Waters technology to quantitate oligonucleotides while providing significant structural information, resulting in both improved quality as well as productivity for biopharmaceutical laboratories, making UPLC/MS an enabling technology for analysis of DNA/RNA based therapeutics.
UPLC analysis of phosphorothioate oligonucleotides: method development. Waters Application Note. 2007: 720002405EN.
UPLC/MS analysis of interfering RNA oligonucleotides. Waters Application Note. 2007: 720002412EN.
720002413, February 2008