100 Samples per Hour: High-Speed Oligonucleotide Analysis with BioAccord™ LC-MS and INTACT Mass Application
Jonathan Fox, Laetitia Denbigh, Scott Berger, Kate Yu
Waters Corporation, United States
Published on November 27, 2025
Abstract
The development of oligonucleotide therapeutics requires fast, reliable analytical workflows to support screening, synthesis, and quality control. This application note presents a streamlined solution using the Waters BioAccord LC-MS System operated under the waters_connect™ Informatics Platform, utilizing the INTACT Mass Application to deliver automated mass confirmation and purity determination of oligonucleotides. By integrating robust LC-MS capabilities with intelligent and automated data processing, the system enables acquisition and analysis of over 100 samples per hour. The automated workflow minimizes manual interventions, enhances the consistency of results and enables scientists to make faster, more informed decisions in early-stage development.
Benefits
Whether screening early candidates or assaying batches of a commercial product, the BioAccord LC-MS System combined with the waters_connect INTACT Mass App (Figure 1) delivers speed, automation, and precision.
1. Fast, scalable, and automated
- This study demonstrates the analysis of over 100 samples per hour with automated workflow integration from acquisition to processed results.
- The automated processing eliminated manual steps associated with defining parameters for peak detection, mass spectral deconvolution, impurity identification, and report generation.
2. Precision that delivers
- Increased deconvoluted mass accuracy was achieved using the chemical formula for each sample to improve the deconvolution model.
- Embedded purity calculations processed optical chromatograms, TICs, or mass spectra, within the INTACT Mass Application to deliver consistent results.
- Targeted impurities were identified, reporting delta masses for novel, non-targeted species, simplifying downstream investigations.
3. Intuitive insights and visualization
- Color-coded dashboards for mass confirmation and purity thresholds simplified sample review, an important element for eliminating a key bottleneck in high throughput analysis.
- The software flags exceptions to the expected results, with grouped failed samples presented for efficient review.
4. Flexible platform, compliance-ready
- The platform enables rapid deployment of validated methods with a single vendor, compliance-ready software environment for data acquisition, data processing, and result reporting.

Introduction
Synthetic oligonucleotides are increasingly recognized as versatile therapeutic agents, offering high specificity and the ability to target previously inaccessible biological pathways. As the scope of oligonucleotide-based therapies expands from highly modified antisense oligonucleotides (ASOs) to siRNAs and sgRNAs, so does the need for robust analytical workflows capable of handling diverse chemistries, modifications, and sample complexity.
Unlike protein-based biotherapeutics, most oligonucleotides are produced via chemical synthesis, allowing for precise control over sequence and structure. However, even with highly optimized synthetic protocols, it remains essential to confirm molecular mass, assess purity, and identify potential failure sequences or synthetic impurities.
This application note presents a higher throughput LC-MS workflow using the Waters BioAccord LC-MS System and INTACT Mass App on the waters_connect Platform, with a workflow designed to automate the routine analysis of synthetic oligonucleotides. The data confirmed successful analysis of oligonucleotides ranging from 10-mer to 60-mer, including ASOs with diverse backbone and sugar modifications. While this range reflects typical therapeutic oligonucleotides, the software can support even greater sequence lengths and structural diversity.
Experimental results demonstrated the system’s ability to deliver rapid, accurate mass confirmation and impurity profiling across a wide oligonucleotide landscape. With acquisition and processing times under 30 seconds per sample, throughput can exceed 100 samples per hour. This facilitates scalable, reliable analysis for screening, development, and quality control of oligonucleotides, empowering researchers to make confident decisions at every stage of analysis.
Experimental
A reversed-phase trap and elute LC-MS method was developed to enable high-throughput intact mass confirmation screening of synthetic oligonucleotides. To demonstrate the method’s versatility, a range of Waters MassPREP™ Oligonucleotide Standards with concentrations ranging from 0.1 mg/ml to 0.01 mg/ml were analyzed alongside representative antisense oligonucleotides (ASOs), including GEM-91 and nusinersen. These examples encompass a diverse array of oligonucleotide chemistries, lengths, and structural complexities. The study highlights the capability of the workflow to acquire and process data for diverse oligonucleotide samples in a single batch, using a single method, and delivering rapid and reliable mass confirmation and impurity results.
LC Conditions
|
LC system: |
Waters ACQUITY Premier UPLC System (Binary) |
|
Detection: |
UV 260 nm |
|
Vials: |
TruView™ LCMS Certified 12 x 32 mm Screw Neck Vial, Total Recovery, with Cap and Preslit PTFE/Silicone Septum (p/n:186005663CV) |
|
Column: |
ACQUITY Premier Oligonucleotide C18 VanGuard™ FIT Cartridge, 130 Å, 1.7 µm 2.1 mm x 5 mm (p/n:186010696) with VanGuard Cartridge Holder (p/n:186007949) |
|
Sample temperature: |
6 °C |
|
Injection volume: |
1 µL |
|
Flow rate: |
1.00 mL/min |
|
Mobile phase A1: |
7 mM triethylamine (TEA) and 40 mM hexafluoro-2-propanol (IonHance™ HFIP Additive (p/n:186010781) in Milli-Q® water (pH 8.6) |
|
Mobile phase B1: |
3.5 mM TEA, 20 mM HFIP in 50% methanol |
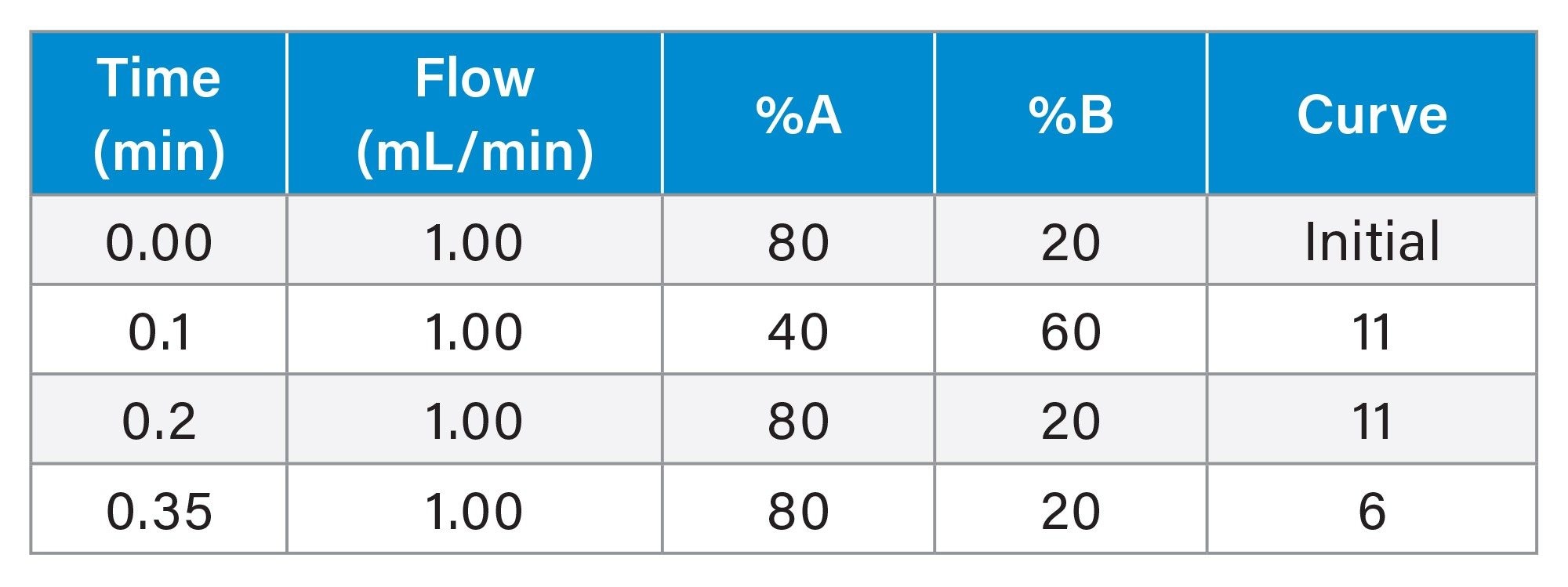
Gradient Table
MS Conditions
|
MS system: |
Waters BioAccord LC-MS System |
|
Mode: |
LC-MSE |
|
Mass range: |
400–5000 m/z |
|
Polarity: |
Negative |
|
Scan rate: |
10 Hz |
|
Cone voltage: |
40 V |
|
Source temperature: |
120 °C |
|
Desolvation temperature: |
550 °C |
|
Capillary voltage: |
0.8 kV |
|
MSE collision energy ramp: |
70–110 eV |
|
Lockmass correction: |
Scheduled lockmass |
The “scheduled lockmass” option was selected in the MS method for this experiment to shorten the total acquisition time; by reducing inter-sample system check activities from a per-injection basis to once every 60 minutes. With the “scheduled lockmass” selected in the INTACT Mass Application, it required only 25 minutes to complete LC-MS data acquisition and data processing for a 48-well tray of oligonucleotide samples.
Results and Discussion
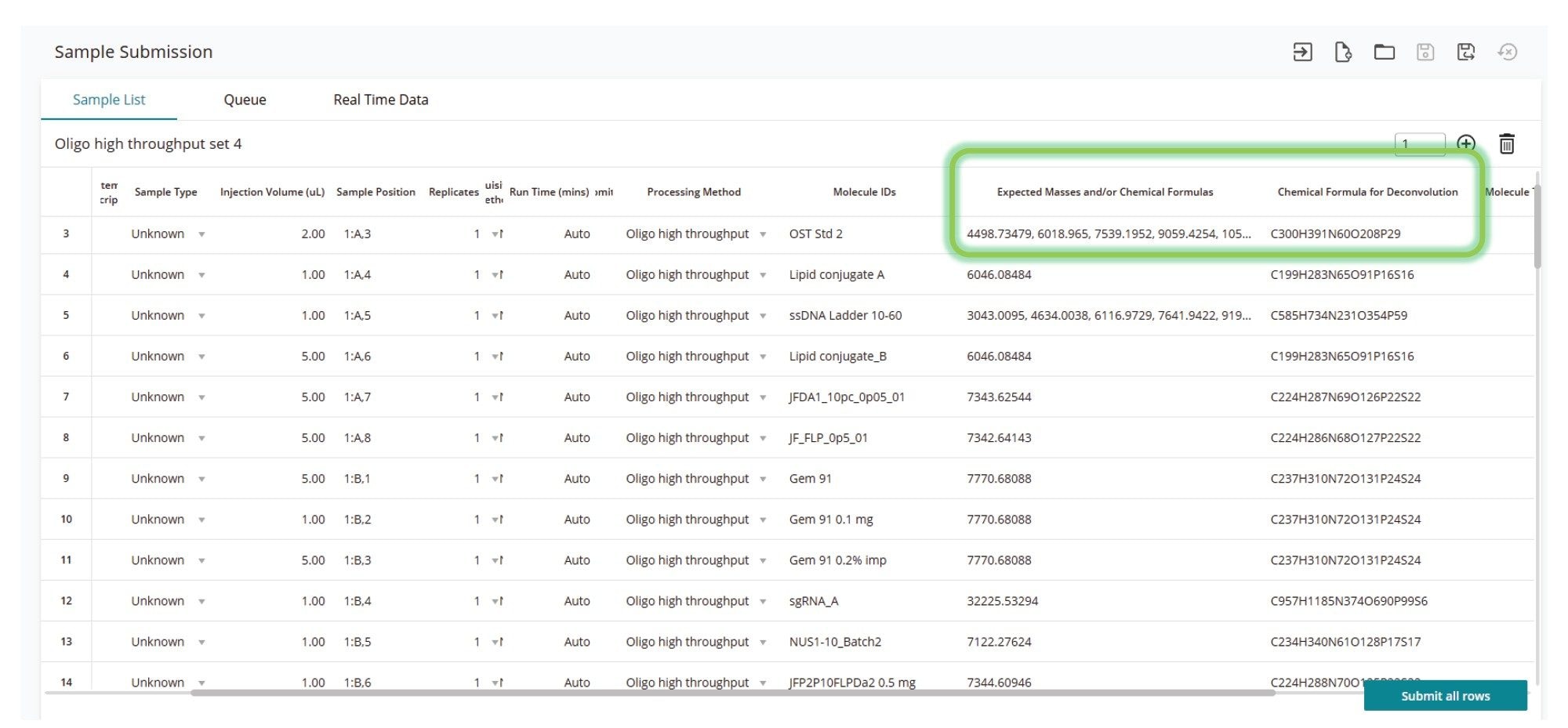
High-throughput analyses present subtle challenges beyond the needs of evaluating a single sample within a batch. Software supporting higher throughput must be flexible enough to deal with analyte diversity within a batch without making methods unnecessarily complex. To address these analytical challenges, the waters_connect INTACT Mass App version 1.9, combined with enhancements to the Sample Submission App 2.7.0, simplifies higher throughput analyses by allowing users to specify either expected masses or chemical formulas directly within the sample list (Figure 2). This significantly simplifies setup, rather than requiring individual methods for each analyte. Additionally, specifying chemical formulas for target molecule assignments enhances the deconvolution model, improving mass accuracy during charge deconvolution. In this study, specifying target chemical formulas enabled successful batch analysis of a diverse range of oligonucleotide samples using a single acquire-and-process method. This approach enabled processing of chemically varied analytes within the same analysis, delivering optimized results for each sample, without the need for manual intervention.
In this study, the BayesSpray deconvolution algorithm with monoisotopic result outputs was used to deliver high precision results for the analyzed oligonucleotide samples.
When analyzing synthetic oligonucleotides, even in higher throughput screening, the analyst is often tasked with assessing critical product attributes beyond simple mass confirmation. This may include detection and reporting of impurities, including known adducts, n–1 modifications, and process-related impurities, recognizing mobile phase adducts as product-related species and reporting unexpected potential impurities by their delta mass from the target molecule.
Supporting this impurity logic within the automated data processing routines reduced the need for manual interventions and can accelerate decision-making, helping scientists move from raw data to actionable insights faster.
Review and Reporting of Results
The dashboard view (Figure 3) provides a real-time summary of oligonucleotide injections during the acquire and process run. Color-coded flags offer instant feedback on each sample’s status, confirming whether target masses were matched and purity thresholds met.
- Green: Pass — mass and purity within expected limits
- Orange: Warning — mass error or reduced purity
- Red: Fail — unmatched mass or purity below threshold
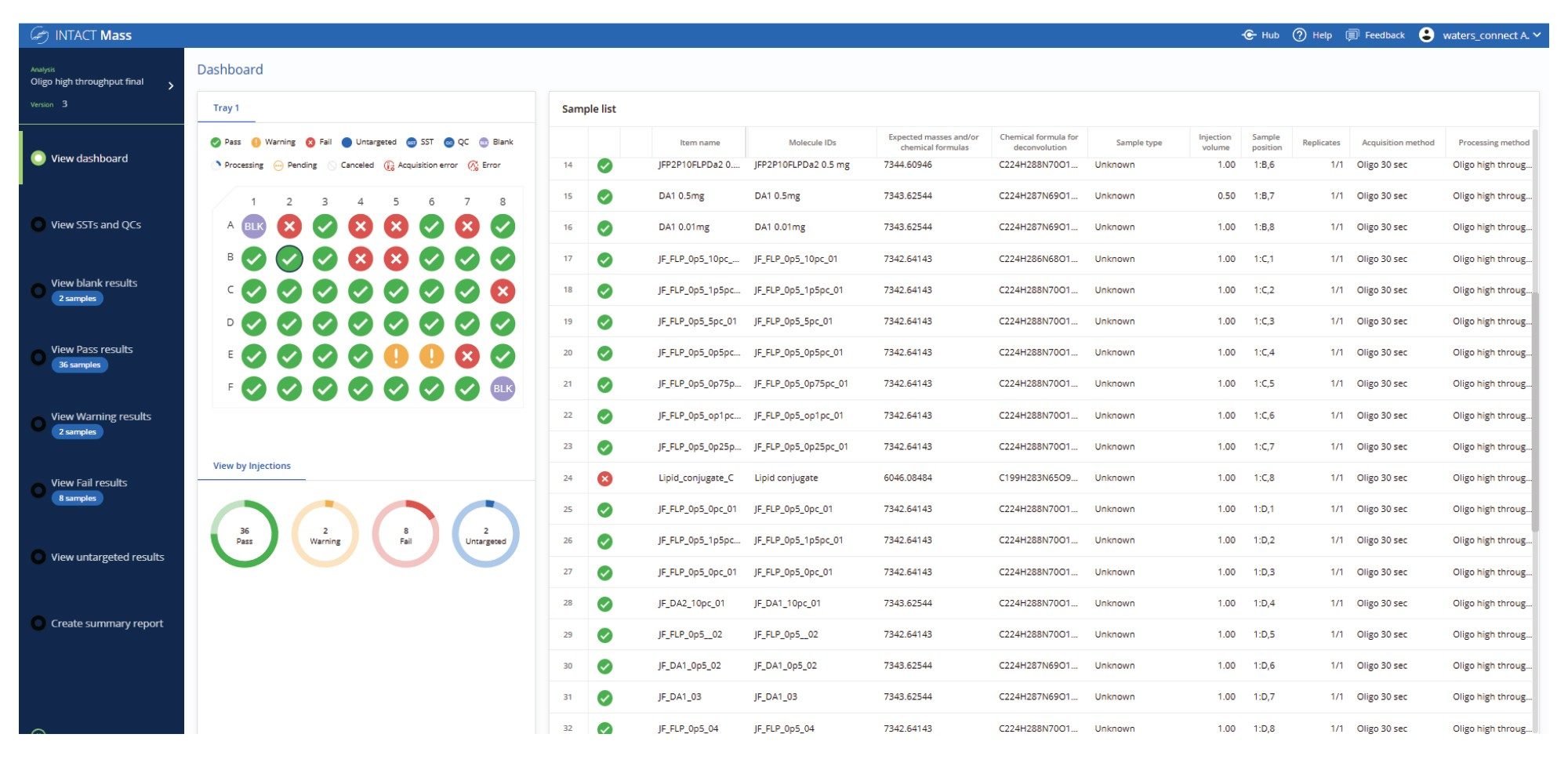
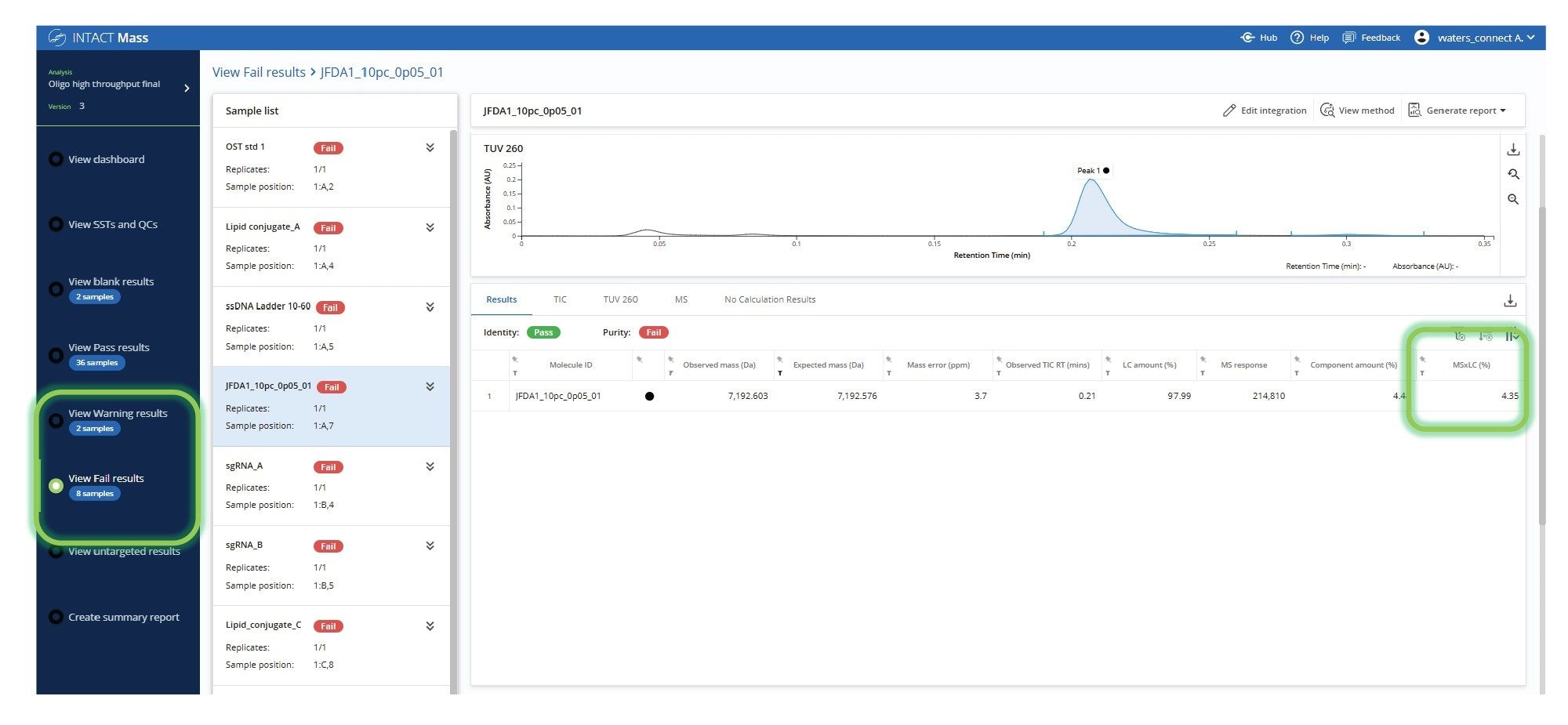
Data review can be a bottleneck in high-throughput analysis, which can be overcome using graphical summaries and flags that quickly communicate the status of each analyzed sample. This is demonstrated in the oligonucleotide plate analysis (Figure 3), where users can distinguish between samples that meet both target and purity criteria, those that fall short of acceptance thresholds, and those flagged for further review. The ability to bring up a grouped display of failed samples (Figure 4) allows the user to quickly focus their review even further and with more detailed results. Samples can be automatically flagged based on key criteria such as mass accuracy, absence of the target mass, or low calculated target purity. These exceptions are clearly visualized and grouped for efficient review, allowing users to quickly focus on problematic data and accelerate decision-making across large batches. In this example, although the target species was successfully identified, the sample was flagged due to not meeting the predefined purity threshold.
To gain deeper insights, results from individual samples can be reviewed individually. Several results spanning a diverse set of oligonucleotides acquired within this batch analysis are provided.
The first result features nusinersen, an oligonucleotide with a phosphorothioate backbone and 2'-O-methoxyethyl (MOE) sugar modifications. The elemental composition (C234H340N61O128P17S17) differs from the typical default deconvolution model settings for oligonucleotides and PS oligonucleotides. By applying the chemical formula, a deconvoluted mass accuracy of -1.5 ppm was achieved for the main target species with no observed artifacts. A range of lower-level impurities and adducts were also identified, with the lowest impurity being quantified at 0.18%. (Figure 5). The INTACT Mass Application method enabled mobile phase adducts to be treated separately from impurities. These adducts can be incorporated into the overall purity percentage of the main product rather than quantified as distinct impurities.

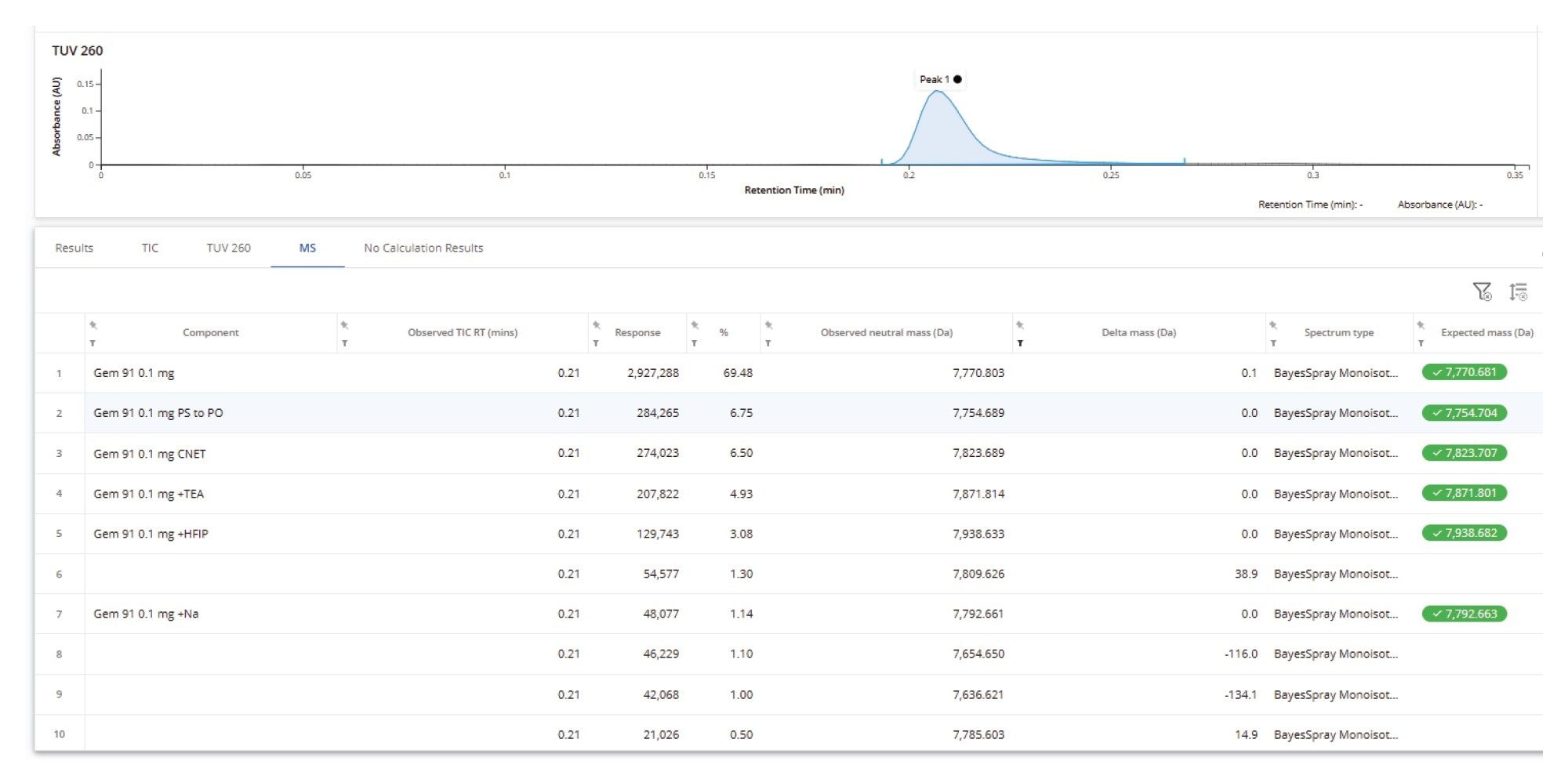
In a separate sample, GEM-91, a fully phosphorothioated 25-mer, was identified with high confidence, and several new impurities were reported. A delta mass calculation is applied to all MS peaks that were not targeted or otherwise identified. This hierarchy of automated assignments streamlines the characterization of novel impurities by highlighting the unexpected species present in the sample.
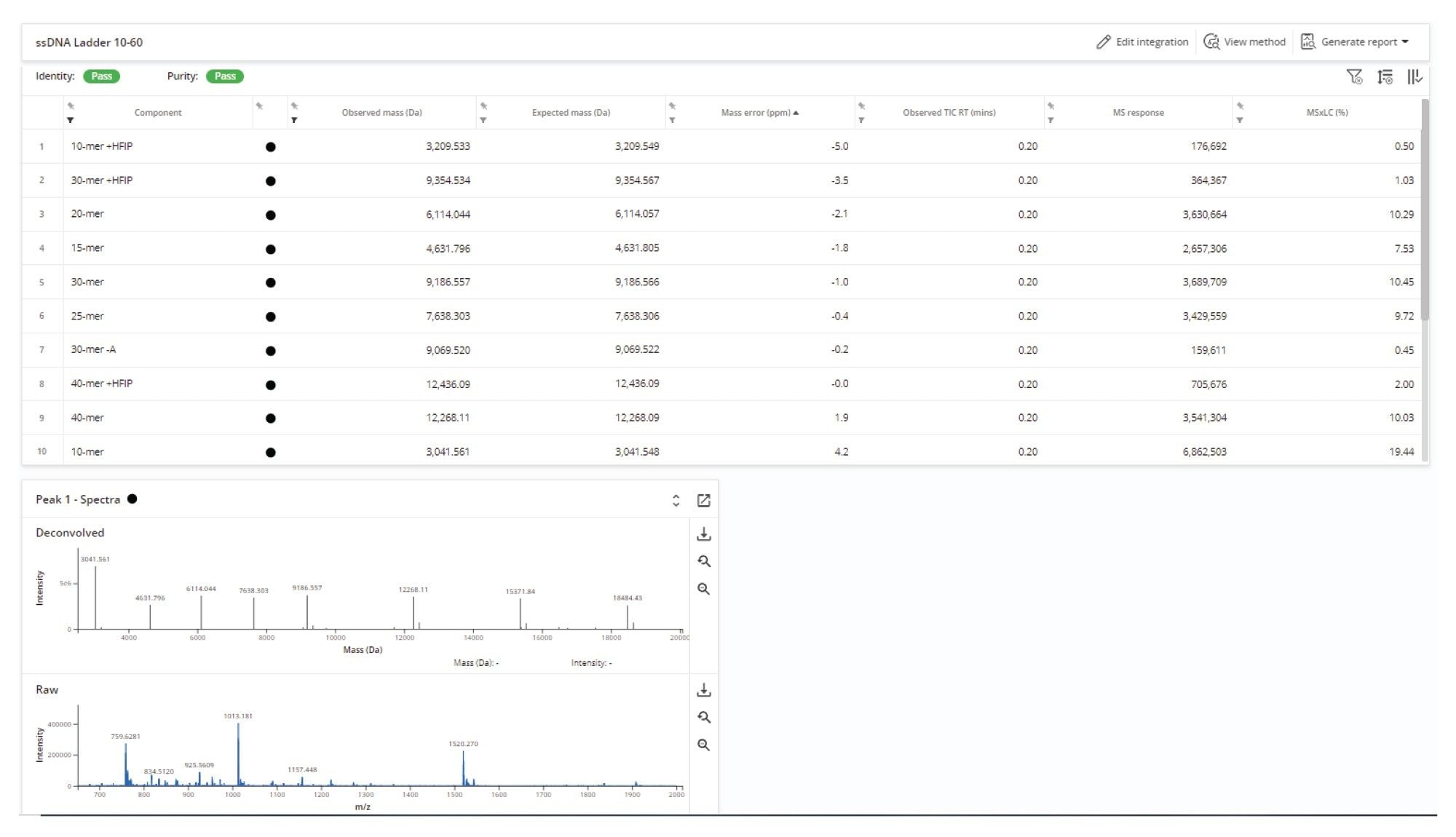
A potential challenge when running batches of samples is the analysis of mixtures containing oligomer products of varying lengths and intensities. However, using the common method for this mixed batch, successful deconvolution and identification of all expected oligonucleotides in the Waters ssDNA ladder was demonstrated, which includes co-eluting species ranging from 10-mer to 60-mer. The lowest abundance species, 10-mer + HFIP, was identified with an MS×LC purity of 0.5% (Figure 7).
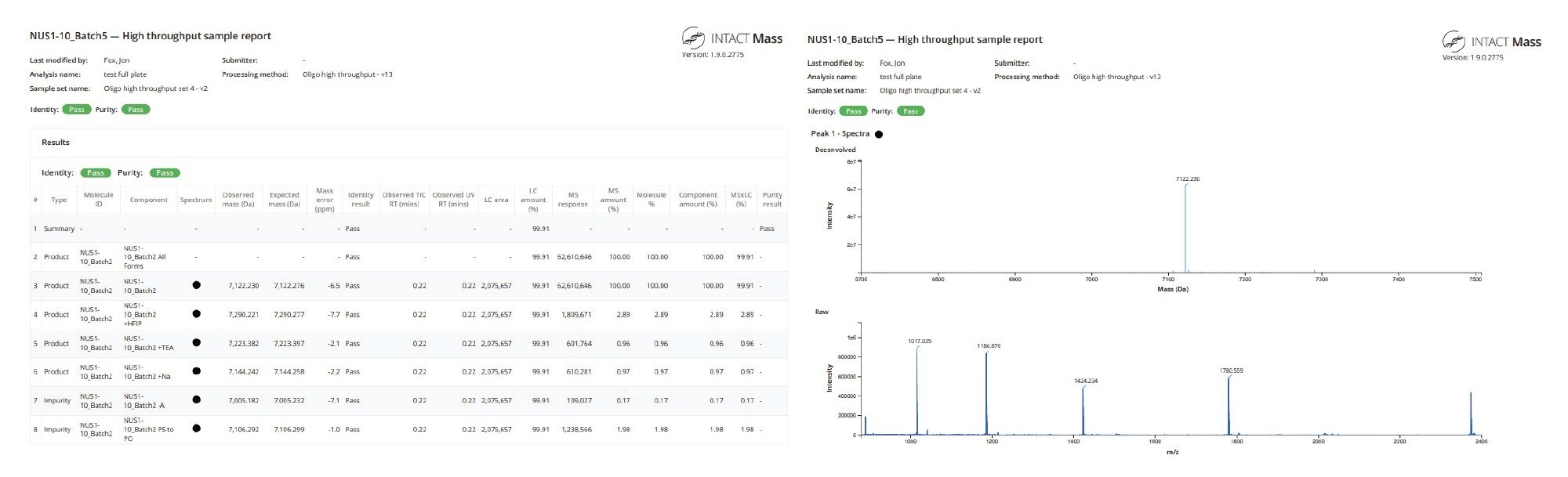
Reporting
Automated reporting ensures traceability and simplifies documentation for regulatory or internal review. When configuring the acquire and process method in the INTACT Mass App, users can opt to automatically generate a PDF report for each sample (Figure 8).
Conclusion
The BioAccord LC-MS System with the waters_connect INTACT Mass App provided a fast, automated solution for oligonucleotide screening analysis, with several key elements directly contributing to the efficiency and quality of higher throughput screening results:
- Rapid analysis: Over 100 oligonucleotide samples per hour throughput was achieved using a rapid LC-MS method, the scheduled lockmass setting for BioAccord System acquisition, and the Acquire and Process acquisition mode for sample to result automation.
- Automated data processing: Intelligent peak picking, mass spectral deconvolution parameter setting, and embedded impurity profiling logic enabled detailed insights on samples of varying complexity.
- Optimal mass spectral deconvolution: This can be achieved across a sample batch with diverse oligonucleotide types and lengths when using chemical compositions for targeted molecules to optimize the deconvolution model for each sample analyzed.
- Graphical batch summaries and review by exception logic: This enabled faster analyst review of data, and the ability to more quickly gain insights on sample results that do not conform to quality criteria defined in the method.
References
- Shion, H., Berger, S. J., Yu, Y. Q. (2020). Application of a Mass Confirmation Workflow for Biotherapeutics Screening. Waters Application Note. 720007027, November 2020
- Shion, H., Boyce, P., Berger, S. J., Yu, Y. Q. (2022). INTACT Mass™ - a Versatile waters_connect™ Application for Rapid Mass Confirmation and Purity Assessment of Biotherapeutics. Waters Application Note. 720007547, February 2022.
Featured Products
720009127, November 2025