This application note aims to describe the process of developing a successful, automated capture and digestion method for hybrid LC-MS/MS quantification of the fusion protein, etanercept.
Prior to quantification, the comparability of the automated and manual preparations was demonstrated in a step wise approach, assessing the critical steps of the protocol individually. This limited the variables during automation optimization and ultimately decreased the time spent on script development. This approach to developing an automated capture and digestion sample preparation facilitated accurate and reproducible quantification of etanercept from rat plasma down to 1 ng/mL.
Automating a hybrid LC-MS/MS sample preparation approach for highly sensitive and reproducible protein quantification allows walk away sample preparation, a task that would normally occupy almost two full days of a scientists’ time.
Automated liquid handlers in bioanalytical laboratories are routinely used to simplify and standardize sample preparation workflows, ultimately increasing throughput, reducing error, and improving assay performance. While used regularly for simple bioanalytical tasks, like serial dilution, protein precipitation, or solid-phase extraction, implementation of the liquid handlers for complex, multi-step workflows, like protein digestions, has not seen the same success. This could be attributed to the complicated method development and optimization of steps like immunoaffinity purification and protein digestion to achieve the high levels of sensitivity desired for accurate quantification from biological matrices. Successful automation implementation of the previously described protein quantification workflow requires assessment, verification, and potential re-optimization of the sub-steps contained within to ensure it meets the rigors of bioanalytical method development criteria.
This application note aims to describe the process of developing a successful, automated capture and digestion method for hybrid LC-MS/MS quantification of the fusion protein, etanercept (Enbrel). Prior to quantification, the comparability of the automated and manual preparations were demonstrated in a step wise approach, assessing the critical steps of the protocol individually. This limited the variables during automation optimization and ultimately decreased the time spent on script development. This approach to developing an automated capture and digestion sample preparation facilitated accurate and reproducible quantification of etanercept from rat plasma down to 1 ng/mL.

a. Plasma sample preparation: Infliximab was spiked into rat plasma at 10,000 ng/mL. A 65 µL aliquot of the prepared infliximab sample was transferred into the Waters 96-well plate and placed on the deck of the STAR (Figure 1).
b. Immunoaffinity reagents: Magne Protein A Beads (Promega, G8781) were used for the purification of infliximab. Tris Buffered Saline (TBS) (25 mM Tris, 150 mM NaCl, pH 7.2) was used as the wash/bind buffer and was poured into the high volume reagent reservoir and placed on deck. A 0.1% formic acid solution was used as the elution solution, while a 500 mM ammonium bicarbonate (pH 8.0) solution was used for sample neutralization. The beads, elution, and neutralization buffers were aliquoted into the low volume reservoir and placed on deck.
c. Immunoaffinity sample purification: The following steps were performed manually or entirely automated by the STAR:
i. For each sample, 25 µL of Protein A bead slurry was aliquoted on top of 200 µL of TBS. The sample plate was mixed for 30 seconds and the beads were allowed to settle a top the 96-well plate magnet for two minutes. The entirety of the supernatant was removed and discarded.
ii. The beads were washed twice; following the same protocol as Section i, with 250 µL of TBS, mixed for 30 seconds, and allowed to settle for two minutes before the supernatant was removed.
iii. To purify the infliximab sample, 200 µL of TBS and 50 µL of spiked or blank plasma was aliquoted on top of the magnetic beads. The samples were mixed (1300 rpm) for 1 hour at room temperature. Following incubation, the beads were settled and supernatant was removed and discarded.
iv. The beads were washed twice using 200 µL TBS and the beads were settled and supernatant removed.
v. To elute the bound infliximab from the bead, 80 µL of elution solution was added and mixed (1300 rpm) for 10 minutes at room temperature. The sample eluant was removed and transferred to a clean 96-well plate. The samples were neutralized with the addition of 8 µL neutralization buffer.
d. Digestion: The purified samples were immediately digested using the ProteinWorks Auto-eXpress Low 5 Digest Kit and described protocol (see Section IV for details).

a. Plasma sample preparation:
i. Comparison study: Etanercept samples were spiked at 10,000 ng/mL in rat plasma and prepared in quadruplicate. Infliximab and trastuzumab were used as internal standards (IS) and diluted with TBS to 10,000 ng/mL. 65 µL aliquots of the prepared etanercept samples were transferred into a Waters 96-well plate and placed on the deck of the STAR.
ii. Quantification study: Etanercept standards and QCs were spiked into rat plasma at various concentrations ranging from 1.0–10,000 ng/mL. Trastuzumab was used as IS and was diluted to 1,000 ng/mL with TBS. Both standards and QCs were prepared in triplicate. 65 µL aliquots of the prepared etanercept samples was transferred into the Waters 96-well plate and placed on the deck of the STAR.
b. Immunoaffinity reagents: Goat Anti-Human Biotinylated IgG antibody (Promega, V7830) was coupled with High Capacity Magne Streptavidin Beads (Promega, V7820) for immunopurification of etanercept and IS. The goat anti-human IgG antibody, elution solution (0.1% formic acid), and neutralization buffer (500 mM ammonium bicarbonate, pH 8.0) were placed in the low volume reagent reservoir. The TBS solution was placed into a high volume reagent reservoir and all reservoirs were placed on the STAR deck.
c. Immunoaffinity sample purification: For the Comparison Study, the following steps were performed manually or entirely automated by the STAR, while the etanercept Quantification Study was entirely automated by the STAR:
i. Bead charging:
1. For each sample, 25 µL of streptavidin magnetic bead slurry was aliquoted on top of 200 µL of TBS in the 96-well plate. The sample plate was then mixed for 30 seconds and the beads were allowed to settle a top the 96-well magnet for two minutes. The entirety of the supernatant was removed and discarded.
2. The beads were washed twice; following the same protocol as Section i, with 250 µL of TBS, mixed for 30 seconds, and allowed to settle for two minutes before the supernatant was removed.
3. To charge the beads, a 50 uL aliquot of the biotinylated goat anti-human IgG antibody was added to the beads. The samples were mixed (1300 rpm) for two hours at room temperature.
4. Following charging, the samples were diluted with 200 uL TBS, mixed (1300 rpm), and settled for two minutes before the supernatant was removed.
5. The beads were washed twice; following the same protocol as Section i, with 200 µL of TBS, mixed for 30 seconds, and allowed to settle for two minutes before the supernatant was removed.
ii. Immunoaffinity Sample Purification:
1. To purify the sample, 200 µL of TBS, 50 µL IS, and 50 µL of etanercept spiked or blank plasma was aliquoted on top of the charged magnetic beads.
2. The sample and beads were mixed (1300 rpm) at room temperature overnight. Following incubation, the beads were settled and the supernatant was removed and discarded.
3. The beads were washed twice using 200 µL of TBS and the supernatant removed.
4. To elute the bound etanercept and IS from the bead, 80 µL of elution solution was added and mixed (1300rpm) for 10 minutes at room temperature. The sample eluant was removed and transferred to a clean 96-well plate and neutralized with the addition of 8 µL neutralization buffer.
d. Digestion: The purified samples were immediately digested using the ProteinWorks Auto-eXpress Low 5 Digest Kit and described protocol (see Section IV for details).
a. Protein digestion for Protein A method development (Section II): The entirety of the purified supernatant (≈88 µL) was digested using ProteinWorks Auto-eXpress Low 5 Digestion kit (p/n: 176004078) and provided low volume protocol. The Low 5 digestion protocol includes: denaturation, reduction, alkylation, digestion, and quench steps. This digestion protocol was performed manually.
b. Protein digestion for goat anti-human method development (Section III.a): The entirety of the purified supernatant (≈88 µL) was digested using ProteinWorks Auto-eXpress Low 5 Digestion kit and provided low volume protocol. The Low 5 digestion protocol includes: denaturation, reduction, alkylation, digestion, and quench steps. This digestion protocol was performed manually.
c. Protein digestion for quantification assay of goat anti-human (Section III.b): The entirety of the purified supernatant (≈88 µL) was digested using ProteinWorks Auto-eXpress Low 5 Digestion kit and provided low volume protocol. The Low 5 digestion protocol includes: denaturation, reduction, alkylation, digestion, and quench steps. This digestion protocol was performed by the Hamilton STAR. Comparison of the automated and manual digestion can be seen in the Waters Application Note 720006208EN and performance of the automated script can be seen in the Waters application notes 720006165EN and 720006209EN.
|
LC system: |
ACQUITY UPLC I-Class Plus |
|
Detection: |
Mass Spectrometer |
|
Column: |
ACQUITY UPLC Peptide BEH C18 Column 300Å., 1.7 μm, 2.1 x 150 mm |
|
Column temp.: |
55 °C |
|
Sample temp.: |
10 °C |
|
Injection volume: |
10 μL |
|
Flow rate: |
0.300 mL/min |
|
Mobile phase A: |
0.1% formic acid in water |
|
Mobile phase B: |
0.1% formic acid in acetonitrile |
|
Gradient: |
2–40% mobile phase B in 7.5 minutes |
|
MS system: |
Xevo TQ-XS Mass Spectrometer |
|
Ionization mode: |
ESI+ |
|
Acquisition range: |
MRM |
|
Capillary voltage: |
3 |
|
Cone voltage: |
35 |
|
Chromatography and MS Software: |
MassLynx |
|
Quantification Software: |
TargetLynx |
Fusion proteins are categorized under the biologics market which is anticipated to reach USD 399.5 billion by 2025.2 Though fusion proteins are only a fraction of this pharmaceutical market (largely owned by monoclonal antibodies), one of the top five best selling drugs in 2017 was the fusion protein etanercept with a global revenue of USD 7.98 billion.3 With continued growth of large molecule biotherapies, the need to develop methods which can accurately quantify them from biological matrix in support of drug discovery and research will continue to increase. For protein quantification via LC-MS, this typically includes an enzymatic digestion to break down the proteins into smaller peptide fragments. With many steps and various reagents, method development of the entire process can be time consuming and complex. Adding to this complexity, immunoaffinity purification prior to digestion is often required to improve selectivity and sensitivity.
Identification and optimization of surrogate, tryptic peptides are critical aspects needed for successful MS method development. Etanercept (Figure 2) is particularly difficult to quantify via the surrogate peptide approach due to its high abundance of N- and O-glycans. In order to measure a glycosylated peptide, or glycopeptide, the mass of the glycan would have to be considered in the masses of the MRM transition, or the glycans would have to be removed enzymatically. Since the glycosylation sites of etanercept are well documented, Skyline
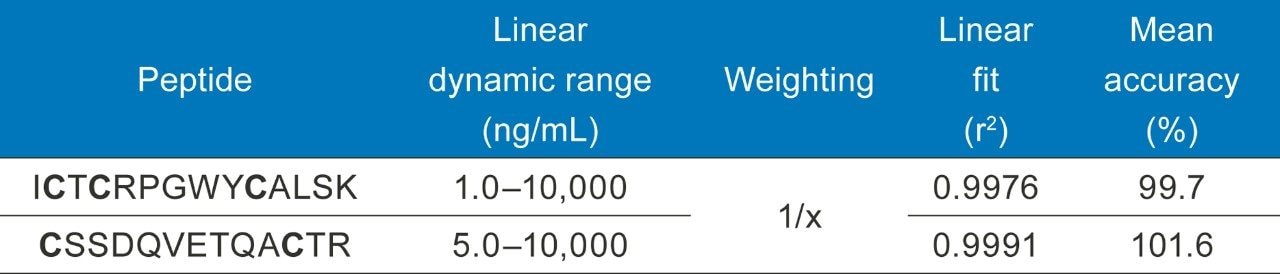
(MacCoss Labs, University of Washington)4 was used for in-silico digestion to determine if any non-glycosylated surrogate peptides exist. Following identification of two non-glycosylated peptides, development and optimizations of the MRM method was experimentally determined using a Skyline/MassLynx workflow performed on a Xevo TQ-XS MS using a tryptic digest of etanercept in buffer. MRM development for infliximab and trastuzumab followed the same process of etanercept, using Skyline and MassLynx for optimization. The amino acid sequence of etanercept is shown in Figure 2, with the surrogate peptides used for quantification highlighted in green. Optimized MS conditions and MRM transitions for etanercept tryptic peptides are listed in Table 1.

Chromatographic separation of etanercept tryptic peptides was achieved using an ACQUITY UPLC Peptide BEH C18, 300 Å, 1.7 µm, 2.1 × 150 mm Column. A shallow gradient from two to 40% mobile phase B over 7.5 minutes afforded the best chromatographic performance, ensuring retention of the polar CSSDQVETQACTR (CSS) peptide.
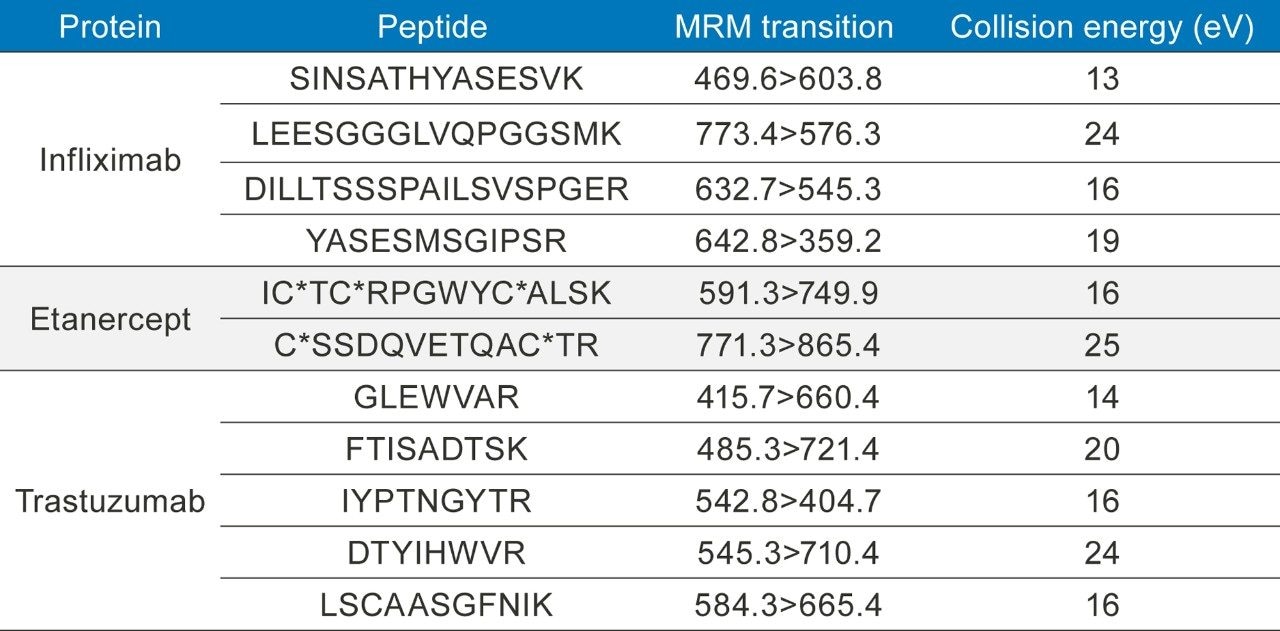
Table 2. MRM conditions for infliximab, etanercept, and trastuzumab, all used in the optimization of automating an immunoaffinity magnetic bead capture.
*Denotes a carbamilation (CAM) of the cysteine residue contributing +57 amu.
One of the first steps of developing a successful automation strategy is to develop the automation script to perform all the tasks of the manual workflow. Water testing should be employed to verify the visual success of various movements and liquid handling. Then its performance should be tested against the manual workflow. A discrete sample set used for both manual and automated strategies should verify the initial performance of the script. Demonstrating the comparability between manual and automated performance ensures what is previously expected of assay performance to be maintained with switching to automation. Once comparable performance is achieved, further experiments must be designed to assess the overall quantitative performance of the automated workflow. In this case, after the automated and manual capture comparison was acceptable within the internally established guidelines (+/- 15% peak area and <20% RSD), a full quantification assessment, using a full set of calibration standards and QCs, was performed.
It is no secret that immunoaffinity purification is expensive. Depending on the specificity of the purification (generic to specific), the cost per sample typically ranges between 4–20 USD. This is a major reason why many scientists decide to implement generic captures like Protein A (less selectivity) over specific captures like goat anti-human from pre-clinical species. Additionally, the development of such a costly and complex product is extremely time consuming and difficult. It is these reasons why the development of the automated script for affinity purification was initially tested with a less expensive, Protein A, generic affinity capture, which has a high binding affinity for human IgG1.
This mechanism works by capturing the human IgG1 component of the drug, then washing away the unbound components, leaving the sample purified of non-specific proteins (Figure 3).5 More importantly, the Protein A magnetic bead capture (Section II.c.) is step for step the same as the purification for Goat Anti-human IgG (Section III.c.ii), making it a reasonable option for affinity automation script development. By successfully automating the Protein A magnetic bead capture, part two of the anti-human capture should be automated successfully.
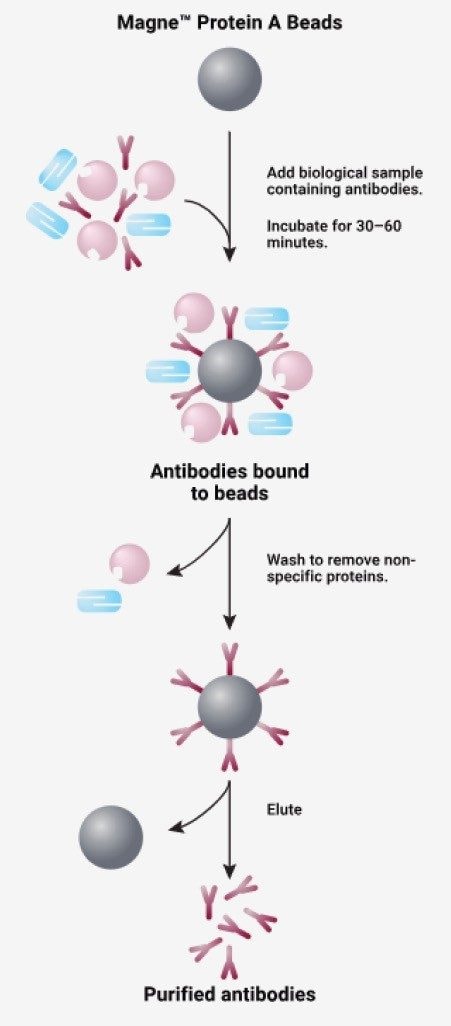
A critical aspect of automating analytical processes is determining the proper labware for execution. Seemingly unimportant for manual work, poor labware or accessory selection could be detrimental for the reproducibility of an automated process. This can make for poor performance and time consuming method development for correction. Compatibility of each should be tested individually before commencing analytical testing.
Prior to automation development, the Protein A magnetic bead purification was optimized manually. This way, only the automation steps would need to be optimized. This method can be found in the Experimental section. Key observations worth noting include:
For these reasons, the Eppendorf Protein Lo Bind 500 µL well plate was chosen for automating the magnetic bead purification sample preparation, offering all of the above requirements for automating this workflow in a 96-well plate format.
Another hurdle of automation is determining the appropriate labware for the application. By looking at each step of the immunoaffinity workflow, the basic components required for automation can be identified. For the immunocapture (generic or specific) of mAbs using magnetic beads, the basic steps include: pipetting, sample mixing, and magnetic bead isolation. For automation of these steps, the liquid handler employed must be configured with:
The final deck layout for the Hamilton STAR performing magnetic bead immunoaffinity purification is shown in Figure 1.
Another difficult step of automating magnetic bead purification is aspirating and dispensing the bead slurry reproducibly. Even without a magnet, the beads will settle quickly to the bottom of any reservoir, making it critically important to re-agitate the beads with pipette mixing to ensure reproducible dispensing to each sample. Otherwise, the concentration of beads will vary between samples causing nonlinear and irregular sample purification. This can be done properly by forcefully aspirating and dispensing back into the reagent reservoir to promote dispersion of the beads throughout the slurry.
Following the optimization of the automated immunoaffinity purification, successful comparison of automated and manual Protein A capture of infliximab was achieved (Figure 4). The area performance and %RSDs are represented well within the internally established acceptance guidelines of +/-15% area of the manual performance and 20% RSD. Since comparable automated performance was achieved, further development of the goat anti-human IgG immunoaffinity purification could begin.
In addition to the capture step of Protein A, the anti-human protocol has an upfront charge step, where the biotinylated goat anti-human IgG antibody is charged to the streptavidin bead, this mechanism can be seen in Figure 5.6 To ensure the success of the charge step, which would mean success of the whole anti-human protocol, each of the wash steps of the purification was mimicked for the charge step, ie. pipette mixing the same volumes, speeds, and heights.
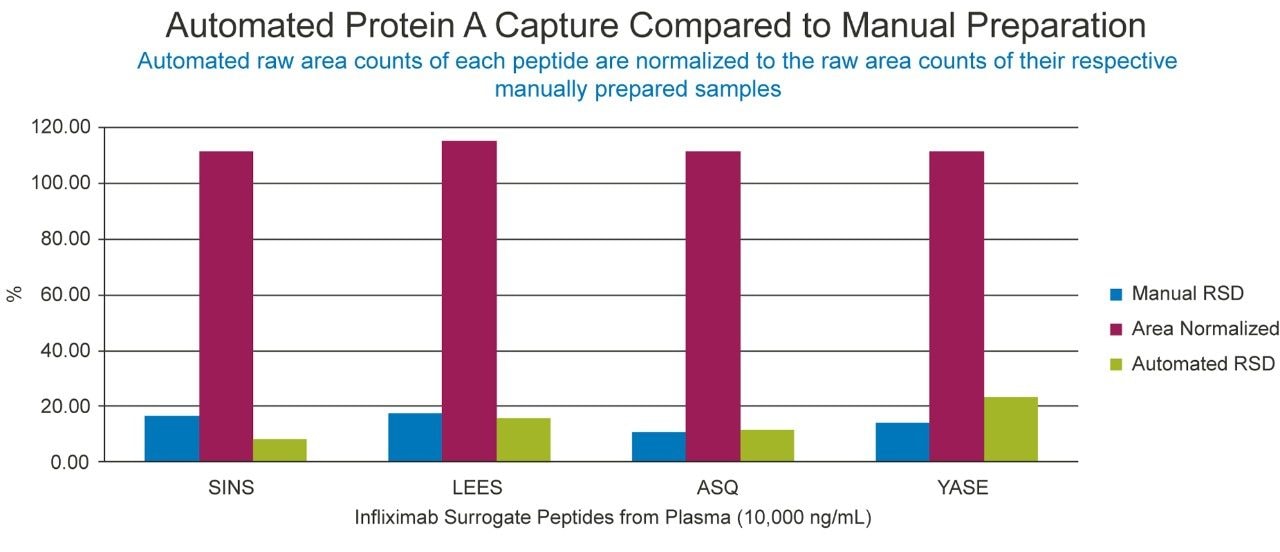
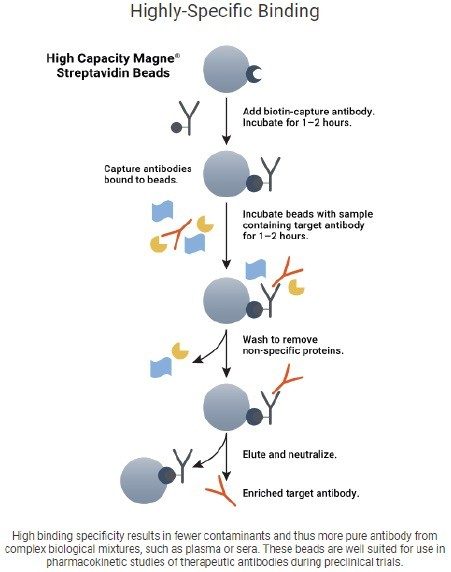
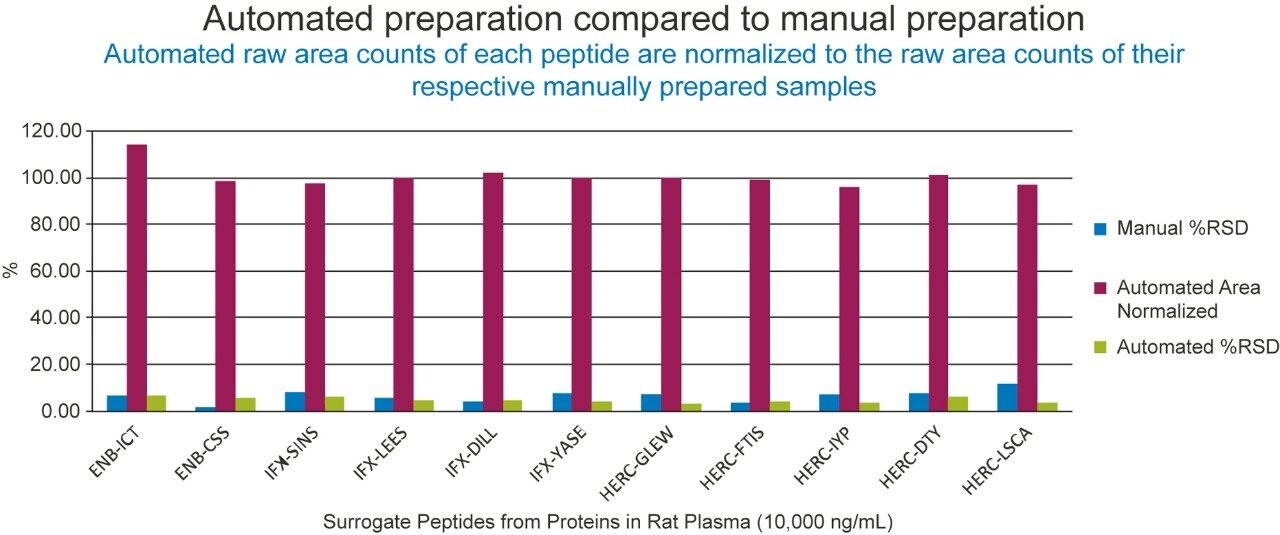
Figure 6 represents the automated and manual comparison of etanercept spiked plasma with infliximab and trastuzumab spiked in TBS as the internal standards. With automated area performance of the anti-human purified etanercept and IS well within 15% of the manual area performance and %RSDs <10% variable, the automated method can be evaluated with a full quantification assay.
Calibration curve standards and QCs were spiked into rat plasma, captured, and then digested on the Hamilton STAR (Section III and IV). Linear dynamic range and quantification statistics for the calibration curve are highlighted in Table 3. The statistics speak to the reproducibility and accuracy of this assay, with linear fits >0.99, QC accuracies within +/-10% (Table 4), and an LOQ of 1 ng/mL, even through multiple complex sample preparation strategies.
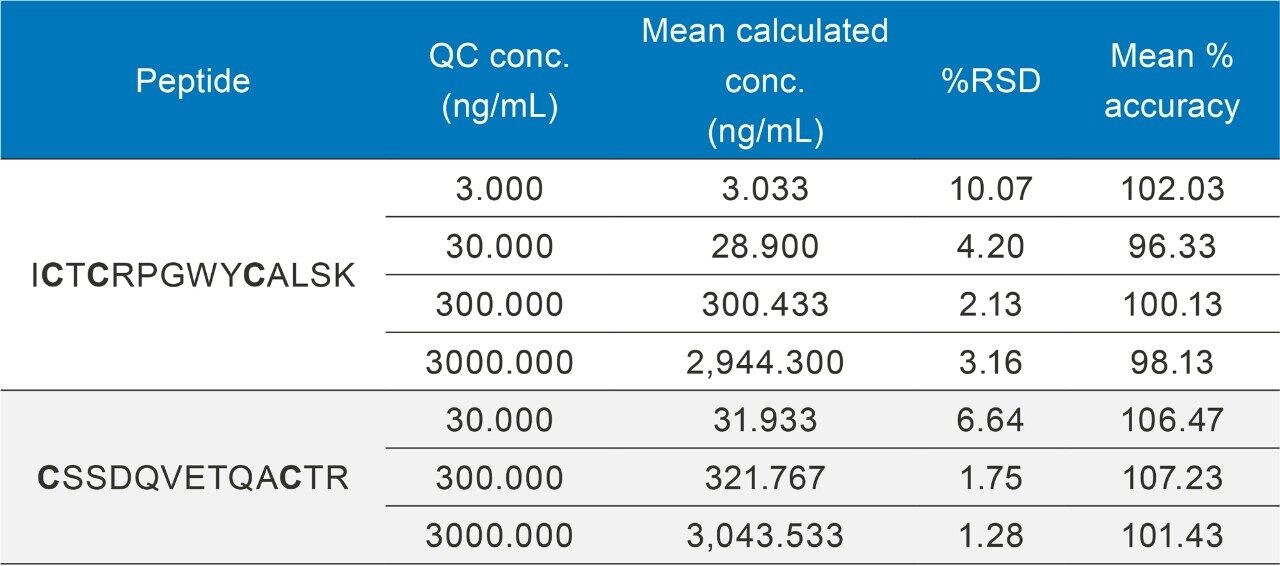
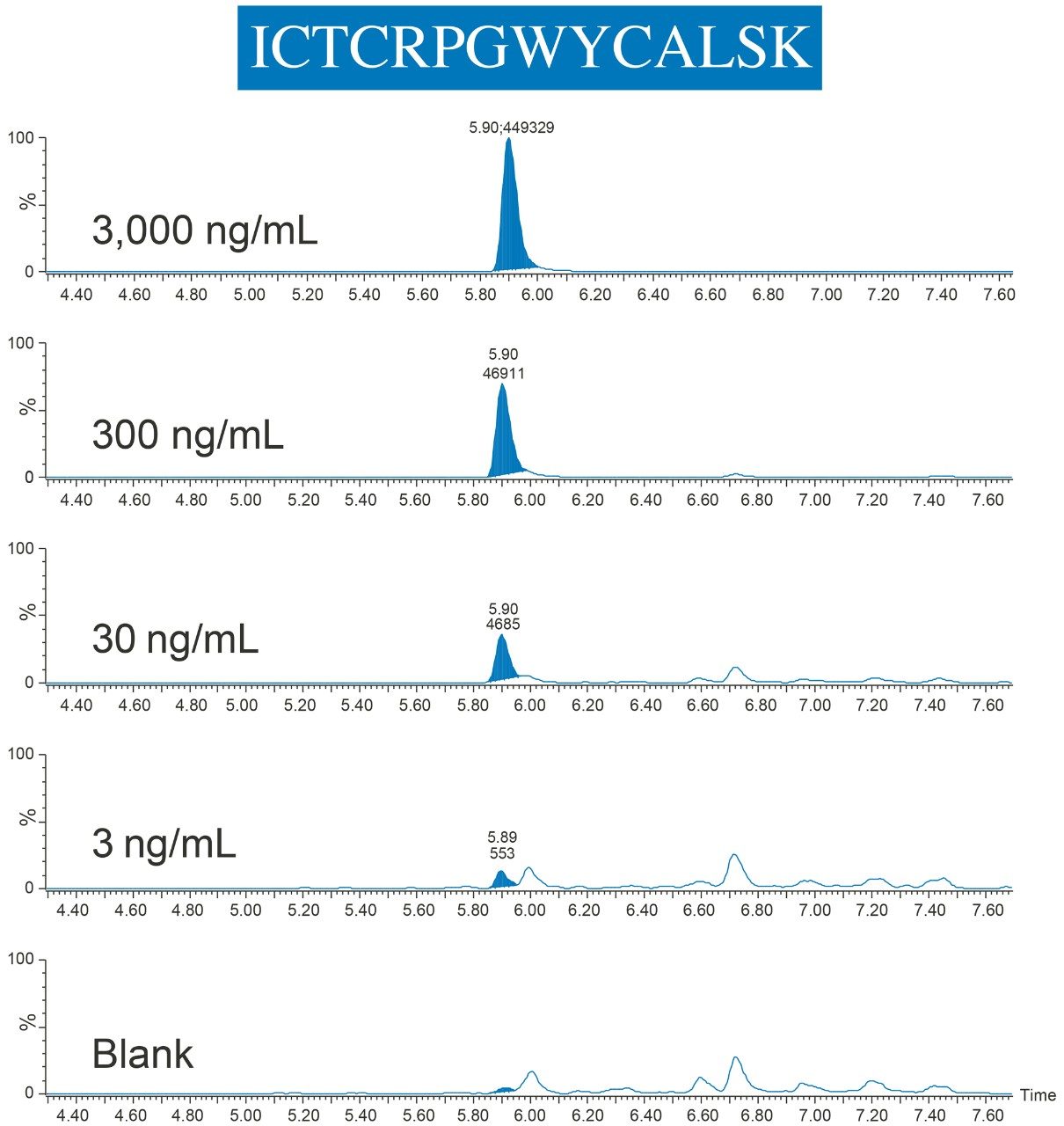
By employing this automated sample preparation approach with affinity capture and enzymatic digestion, this assay met bioanalytical method validation guidelines for precision and accuracy, with QC accuracies between 96–107% and RSDs ≤10%. Example chromatograms of the ICTCRPGWYCALSK (ICT) peptide QCs are illustrated in Figure 7.
Automation has proven to increase lab productivity, reduce human error, and ensure reproducibility between assays. However, for multi-step sample preparation strategies like protein digestion, it can be daunting and time consuming. This is especially true when an upfront immunoaffinity purification of the matrix is necessary to achieve high sensitivity. Taking a step by step approach and assessing critical steps of the process individually can ease the burden during method development. Additionally, using a standardized, kit-based approach optimized for protein digestions reduces the method development of one of the critical steps. Finally, automating a manual process that has been previously optimized can offer a comparison or a starting point for how the method should perform.
Using this strategy for automation script development produced a highly sensitive, accurate, and reproducible hybrid LC-MS/MS method for protein quantification via the surrogate peptide approach. Automating the immunoaffinity purification prior to use of the verified automation script for ProteinWorks Auto-eXpress Digestions on Hamilton STAR systems resulted in robust quantification of etanercept with an LLOQ of 1 ng/mL from rat plasma. If developed properly, automating complex workflows, like protein quantification, can minimize human error, increase throughput and maximize productivity all while achieving accurate and reproducible performance.
720006497, February 2019