For research use only. Not for use in diagnostic procedures.
This is an Application Brief and does not contain a detailed Experimental section.
This application brief demonstrates the application of SONAR on a SYNAPT XS platform for improved specificity, higher sequence coverage, and increased confidence in identification following database search.
The demonstration of SONAR on a SYNAPT XS platform to analyze the plasma proteome.
Traditionally, proteomics has used data dependent (DDA) strategies for mass spectrometry (MS) acquisition. However, developments in data independent (DIA) modes of acquisition have provided the ability to gain qualitative and quantitative information in a single injection without the drawbacks associated with DDA workflows. Several DIA variants exist, but an alternative approach is to apply a DIA methodology that allows for high throughput whilst ensuring high specificity and quantitative accuracy. SONAR has previously been described,1,2 highlighting the utilization of a fast scanning quadrupole, enabling the technique to be compatible with fast chromatography and high-throughput workflows. Here, we present the applicability of SONAR on a SYNAPT XS platform for the identification and relative quantification of plasma proteins from a patient cohort consisting of controls, chronic obstructive pulmonary disease (COPD), and asthma.
The analyzed samples consisted of tryptically digested, undepleted human plasma from three different conditions. Sample groups consisted of controls (n=6), chronic obstructive pulmonary disease (n=6), and asthma (n=6). Individual samples per group were pooled to provide three working samples.
Peptides were separated using an ACQUITY UPLC M-Class System, which was equipped with an ACQUITY UPLC Peptide CSH C18, 300 µm I.D. × 100 mm long analytical column. Samples were analyzed in triplicate (QC=5 injections) based on 5 µg loadings. The samples were separated using a reversed-phase gradient from 1 to 40% acetonitrile (+0.1% formic acid) over 45 minutes at a flow rate of 50 µL/min. Data were collected using a SYNAPT XS which operated using the SONAR mode of acquisition. Data were processed using Progenesis QI for Proteomics and database searched against a human database, consisting of UniProt reviewed sequences. Searches were performed using carbamidomethyl C (fixed) and oxidation of methionine (variable) modifications in addition to a 1% FDR.
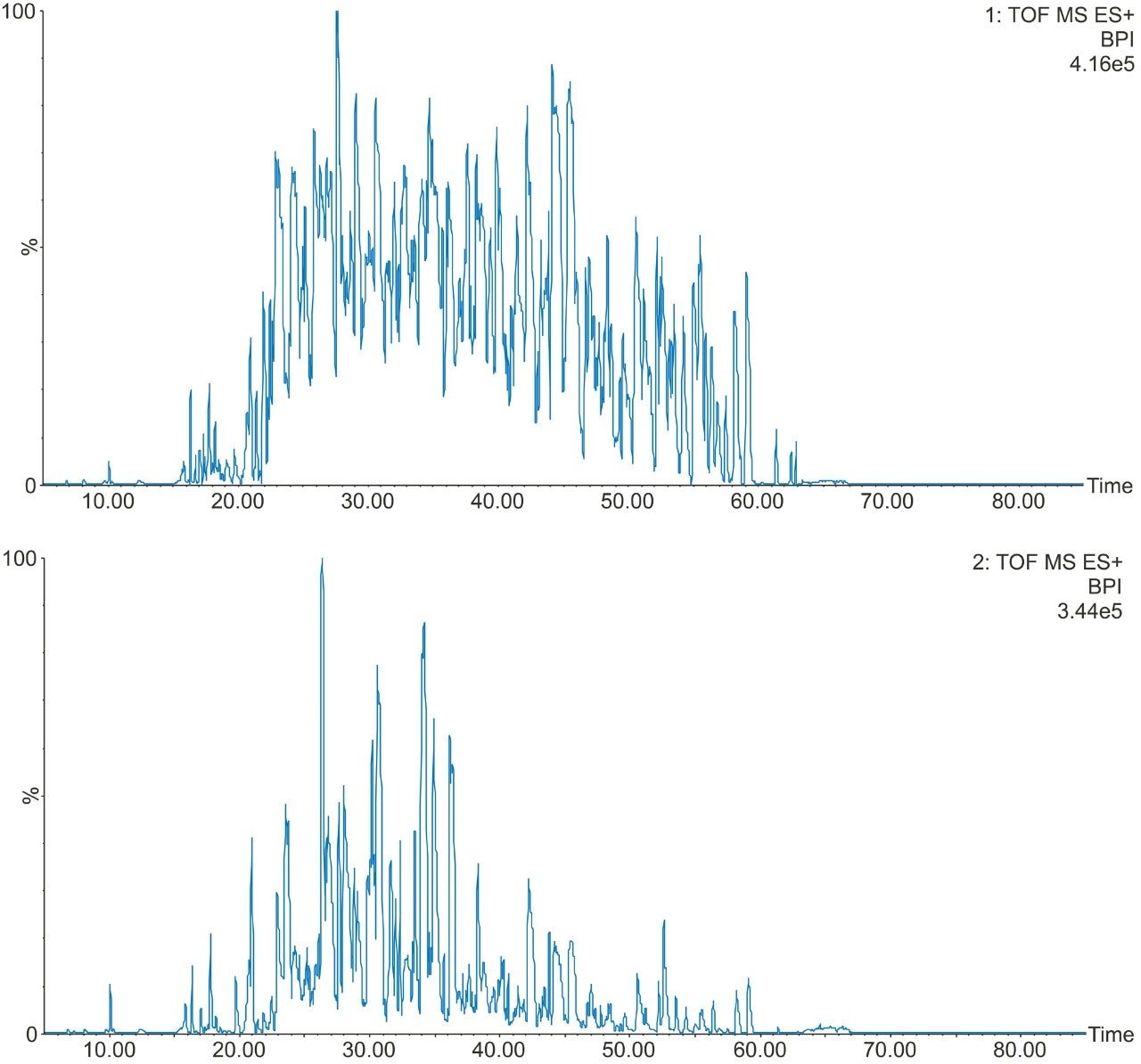
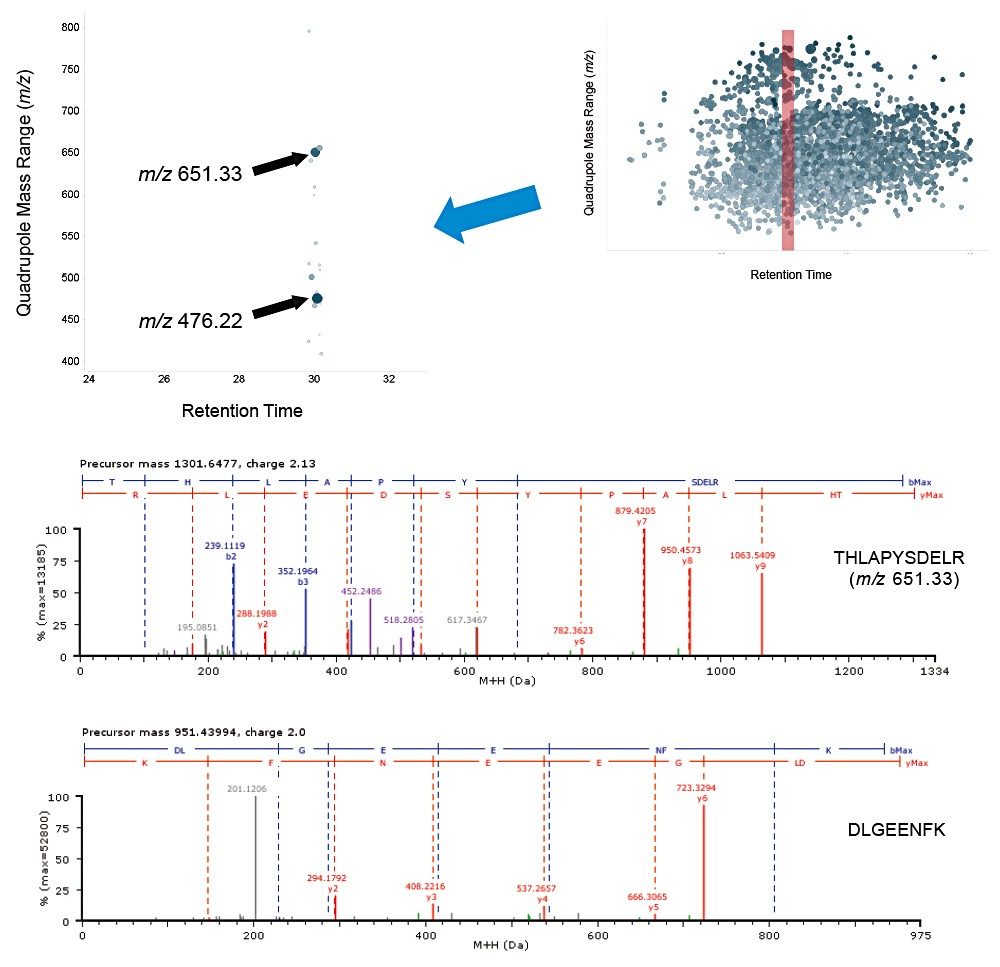
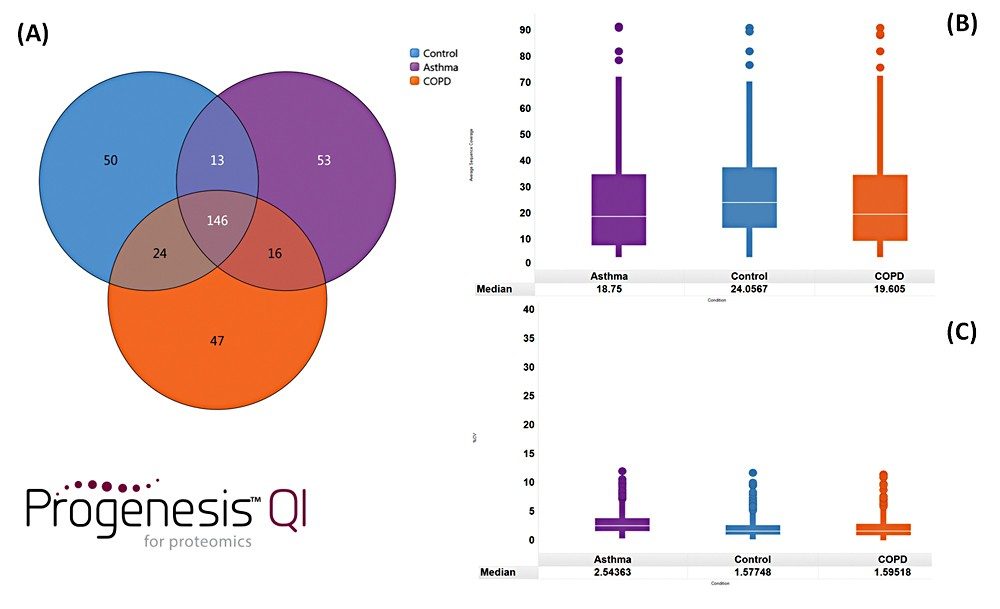
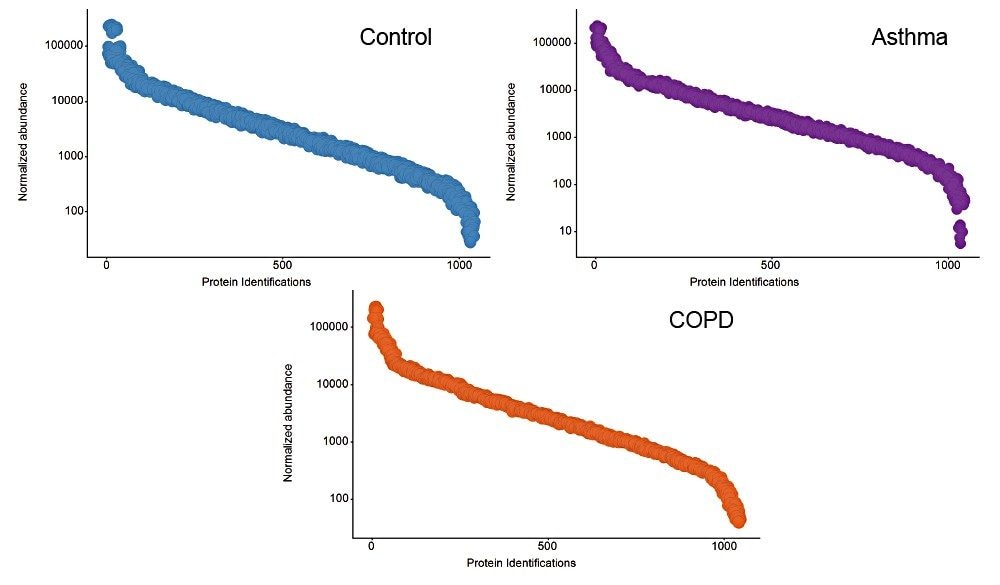
Example chromatograms representing SONAR acquisition are shown in Figure 1 for both precursor and fragment ion data. The benefits of high selectivity when implementing SONAR have previously been reported.1,2 Figure 2 shows the enhanced specificity provided by the scanning quadrupole implemented for increased peptide identification. Data processing with Progenesis QI for Proteomics identified a total of 349 proteins with the majority of identifications overlapping between all three cohorts (Figure 3A), whilst the median sequence coverage achieved varied between 18–25% across groups (Figure 3B). The ability to reproducibly identify and quantify analytes of interest is important to ensure consistency over the analysis. Figure 3C shows the coefficient of variation (CV) for the normalized abundance across technical replicates for each condition, with a median CV less than 3% being achieved across each condition. Furthermore, a linear dynamic range of 3–4 orders magnitude is demonstrated for each condition (Figure 4).
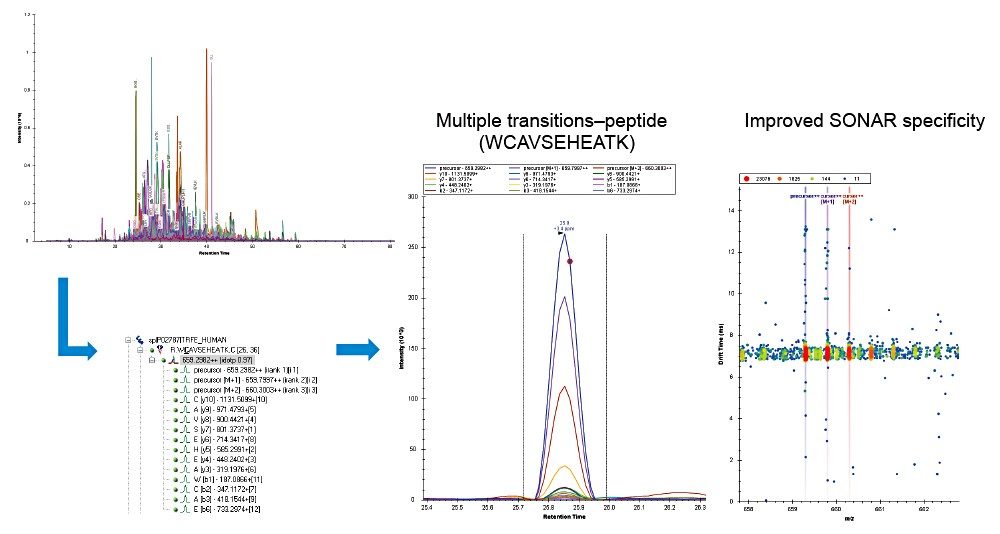
The ability to integrate data with third-party informatics is integral for flexibility and comprehensive data analysis. SONAR generated data is compatible with a variety of software tools and is demonstrated within this study using Skyline software (Figure 5), highlighting serotransferrin as an example protein with associated peptide transitions.
We have shown the utility of SONAR on a SYNAPT XS platform for the proteomic analysis of plasma originating from a respiratory disease patient cohort. Implementing a SONAR acquisition workflow has shown high specificity, wide dynamic range, high protein identification, and sequence coverage. Processing the data through Progenesis QI for Proteomics and other third-party tools, highlights the flexibility of the SONAR workflow. These benefits have been demonstrated with a proteomics example, however the benefits can also be applied to metabolomics and lipidomics for multi-omic-based studies.
720006598, August 2019