This application brief demonstrates to how to enhance selectivity by optimizing quadrupole settings on a quadrupole time-of-flight high resolution mass spectrometer (Q-Tof HRMS) for sensitive quantitation of surrogate peptides in complex matrices.
The development of complex biotherapeutic modalities has led to a need for sensitive and selective quantitative LC-MS techniques that complement and serve as an alternative to ligand binding assays in discovery.
The development of complex biotherapeutic modalities has led to a need for sensitive and selective quantitative LC-MS techniques that complement and serve as an alternative to ligand binding assays in discovery.1,2 High resolution mass spectrometry (HRMS) has certain advantages over tandem quad mass sprectrometry, including high mass resolving power for additional selectivity when faced with a challenging sample and a wider mass range for intact quantification of larger proteins. A single platform for both characterization, as well as targeted modes for quantitation is attractive, providing increased laboratory flexibility.
The surrogate peptide approach is commonly used for protein bioanalysis. Direct digestion of plasma/tissue often results in significant endogenous interference (at both the peptide and fragment level) which can impact both the selectivity and sensitivity of quantitative LC-MS assays for antibody and other protein based drugs. Sample preparation clean-up strategies including immunoaffinity and solid phase extraction are often used to reduce sample complexity and aid in improving selectivity and sensitivity. Optimizing the quadrupole mass selection window on a Q-Tof HRMS can be used to balance the ability to remove matrix interference through selectivity while maximizing sensitivity of a bioanalytical assay.
In this study, quantification of the therapeutic monoclonal antibody, digested trastuzumab, prepared in rat plasma, is demonstrated. Peptide quantitation was performed using HRMS with a Xevo G2-XS QTof which was operated in sensitivity (ESI+) mode and MassLynx Software v4.1. Time-of-flight multiple reaction monitoring (Tof-MRM) was used as the mode of data acquisition.³ The quadrupole mass selection window was varied from 1 Da to 4 Da (LM settings were tuned using leucine encephalin from 4.7 to 15). HM settings were not adjusted. Data was collected from 200 m/z to 1500 m/z and the system calibrated using sodium iodide solution clusters.
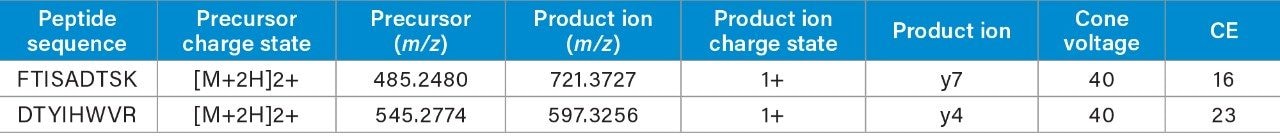
Two unique tryptic peptides of trastuzumab, FTISADTSK and DTYIHWVR analyzed in plasma, were used to demonstrate the benefit of optimizing the quadrupole mass selection window. The m/z of the peptides, their fragments, and corresponding charge states are listed in Table 1.
Chromatographic separation was achieved using an ACQUITY UPLC H-Class System with an ACQUITY UPLC Peptide BEH C18 Column (P/N 186003687), and a gradient (5–50% B) over eight minutes with 0.1% formic acid in both water and acetonitrile (flow rate 0.3 mL/min). Briefly, trastuzumab was purified from rat plasma (50 µL) using affinity capture magnetic Protein A beads.⁴ The post affinity purified plasma was then digested and peptide-level purification was completed using the ProteinWorks eXpress Digest Kit (P/N 176003689) and a ProteinWorks µElution SPE Clean-up Kit (P/N 186008304). A 10 µL aliquot of the resulting 90 µL SPE eluate was injected for each LC-MS analysis.³ The data was processed using TargetLynx Application Manager with a mass extraction window of 50 mDa.
During the digestion process, hundreds of peptides are formed, interference from these peptides is to be expected. When matrix interference occurs close to the target peptide fragment, it can result in poor selectivity and sensitivity. The quadrupole mass selection window can be optimized to either increase transmission of the precursor in relatively clean samples or reduce the transmission of closely eluting peptides of similar mass that generate fragments with mass-to-charge ratios in close proximity or isobaric to the target peptide fragments as is shown in Figure 1. In this example, optimizing the quadrupole mass selection window improved sensitivity and S/N at the low end of the curve by 4 to 8 fold. Although counts were reduced, the decrease in matrix interference significantly improved the results.
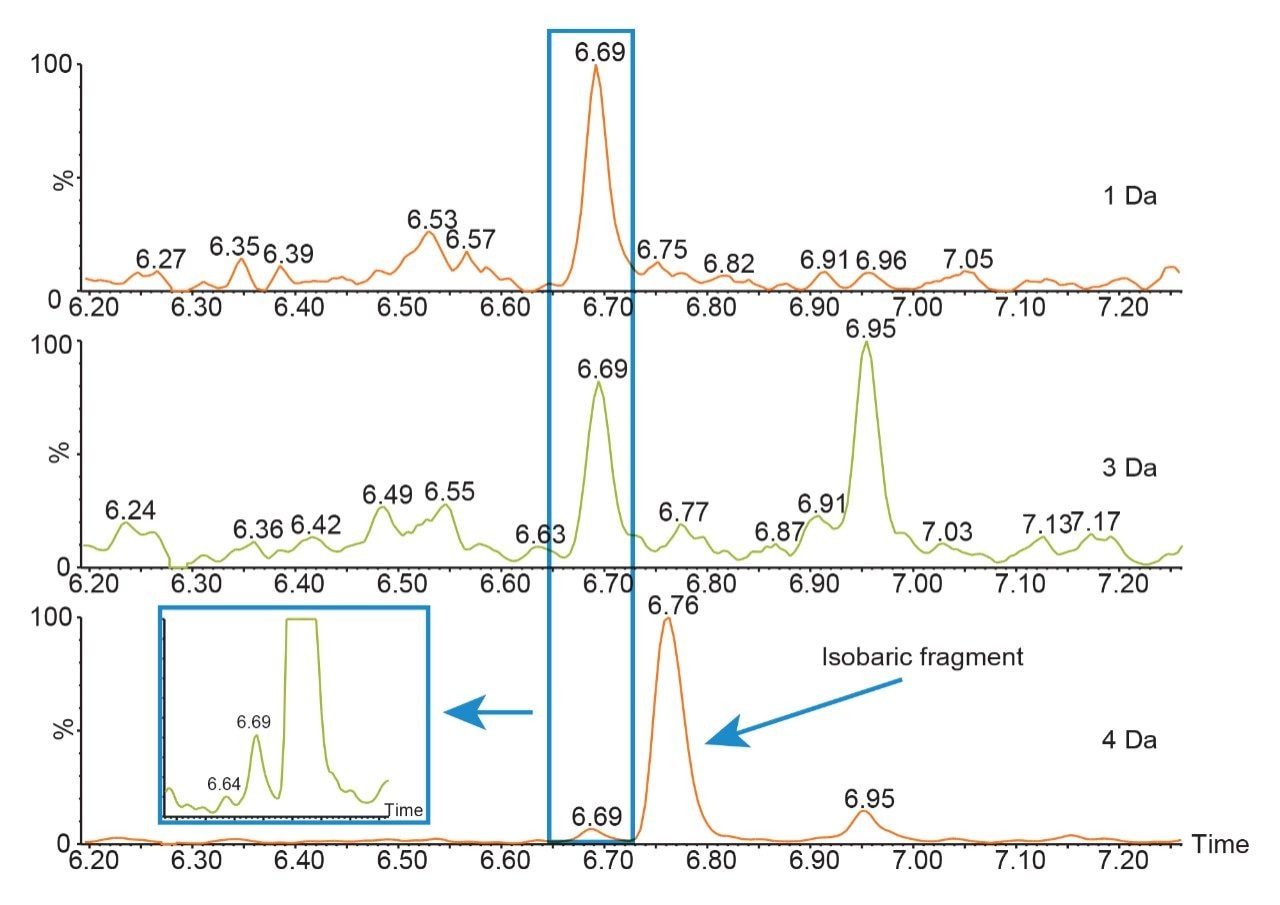
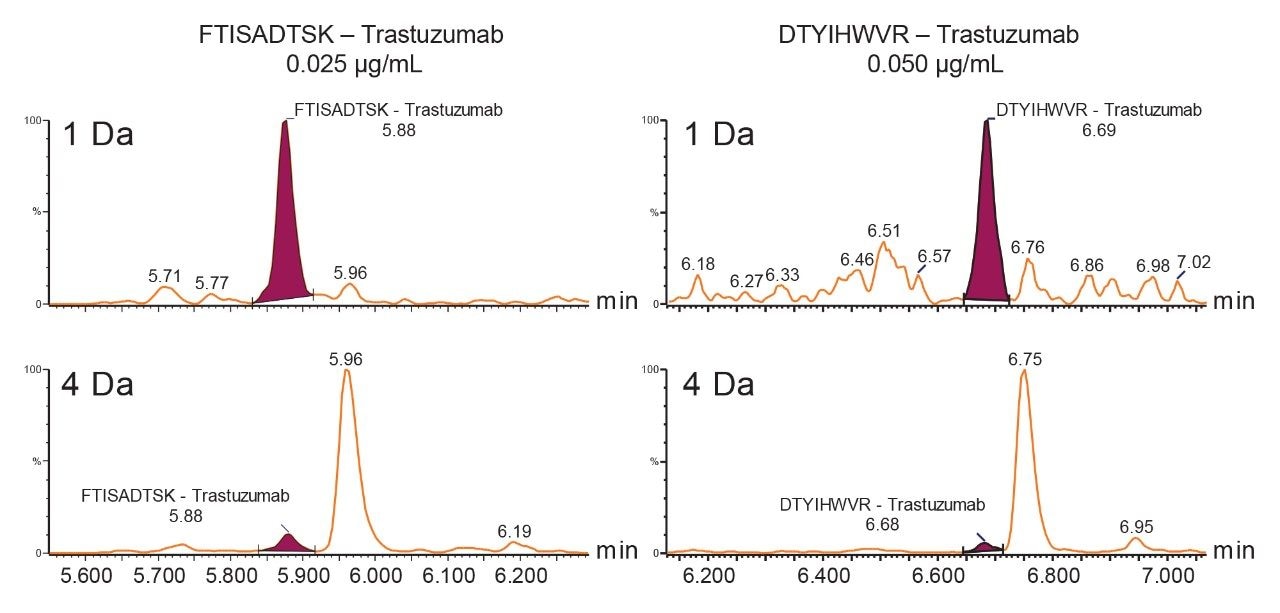
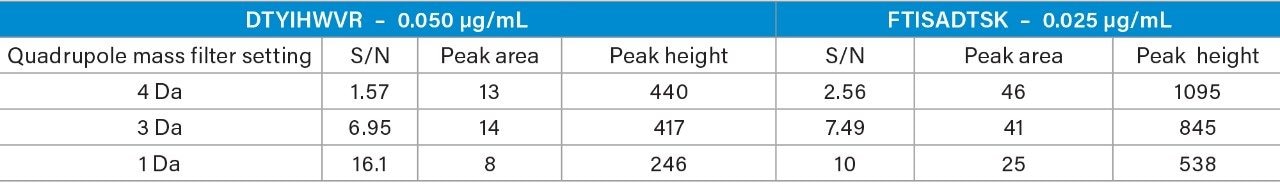
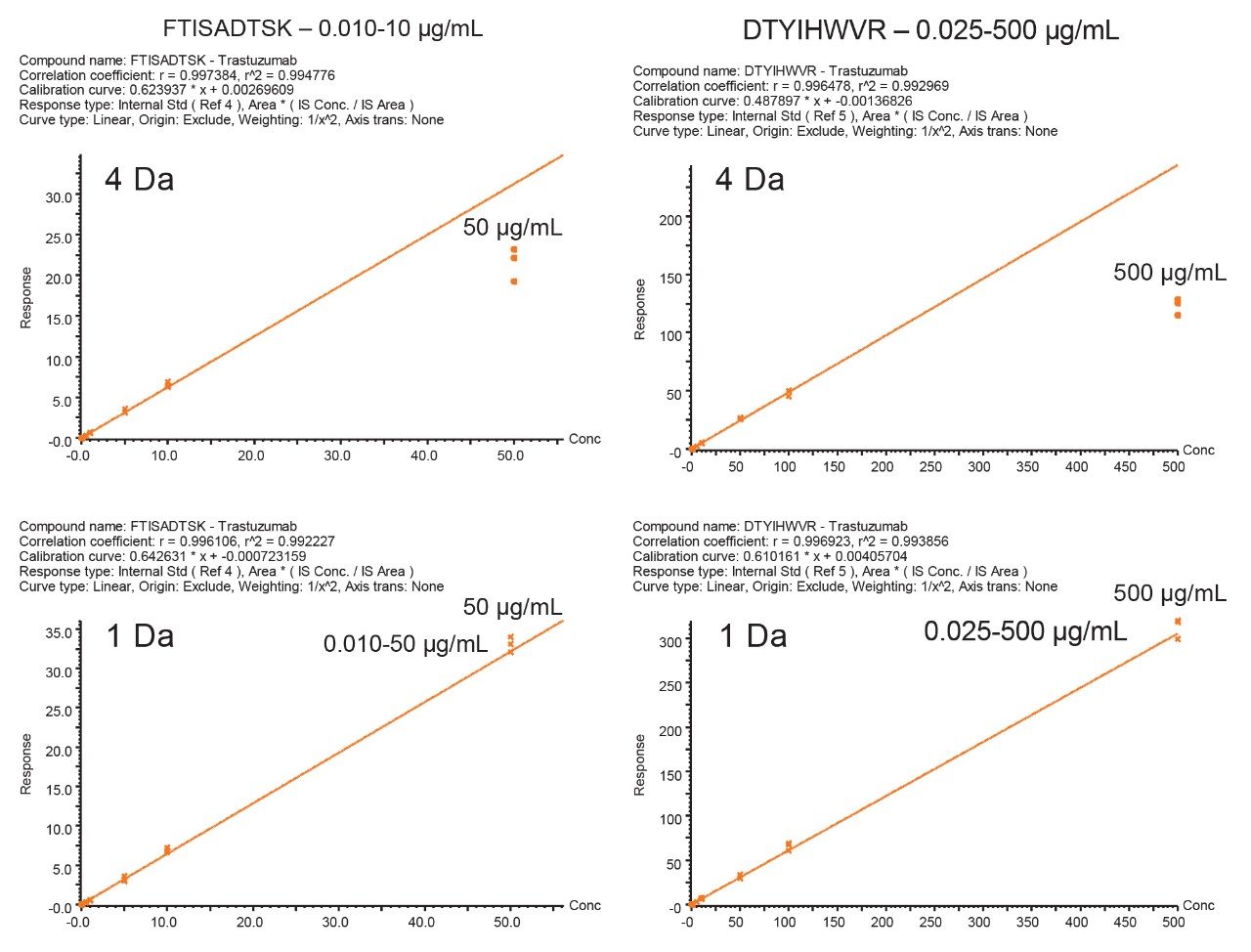
When the quadrupole mass selection window is set to transmit a 4 Da window, matrix interference, which produces an isobaric fragment, elutes in close proximity to the target peptide (Figure 1). The presence of this prominent matrix interference can hinder automatic peak integration across the calibration range. The 3 Da window readily removes the largest interfering peak and might be suitable for some assays. In this example, we preferred 1 Da to maximize the robustness of the assay and remove as much background interference as possible. Setting up the quadrupole to transmit a 1 Da window has the effect of significantly decreasing the intensity of the interference and increasing selectivity and sensitivity (Figure 1). The absolute intensity of the target peptide fragments decreases, however, the signal-to-noise ratio (S/N) at the LLOQ for DTYIHWVR (0.05 µg/mL, 10 µL injection) and FTISADTSK (0.025 µg/mL, 10 µL injection) in this matrix improves. Table 2 shows the effect of narrowing the quadrupole mass selection window and its relationship to peak area, peak height and signal-to-noise (S/N). In this example, changing the quadrupole mass selection window also resulted in a clear increase in dynamic range for both peptides. The calibration plots also revealed that each peptide gained half an order of magnitude using this approach (Figure 3).
The presence of closely eluting matrix peaks can often complicate automatic peak integration of the target analyte. In this example, due to the removal of the interfering peaks, the data processing was simplified leading to more efficient data analysis and reporting.
Isobaric (or close mass to charge) interferences can occur during the analysis of peptides in complex matrices such as plasma/serum and should be investigated during method development. This can impact both the selectivity and sensitivity of quantitative LC-MS assays for antibody and other protein based drugs. Sample preparation, and MS optimization including an optimal quadrupole mass selection window should be used to reduce potential matrix interferences that are close in mass and chromatographic retention time to the peptide and fragment of interest to maximize sensitivity and assay robustness.
720006444, December 2018