The Waters Omics Research Platform with TransOmics Informatics, featuring UPLC and HDMSE technologies, enables researchers to improve how they screen and differentiate molecular phenotypes of plants exposed to different environmental stimuli. This highthroughput approach has applications in agricultural, food, and nutritional, as well as natural product research.
Use of the Waters Omics Research Platform, which combines UPLC with ion mobility LC-MSE and TransOmics Informatics, allows for the rapid characterization of molecular phenotypes of plants that are exposed to different environmental stimuli.
Cruciferous vegetables, such as broccoli, cabbage, kale, and brussels sprouts, are known to be anti-carcinogenic and possess antioxidant effects. They are widely consumed in the world, and represent a rich source of bioactive metabolites.1 Young broccoli plants are an especially good source of chemoprotective metabolites, with levels several times greater than mature plants. Growth conditions and environmental stresses exert a significant influence on the metabolism of broccoli sprouts.2
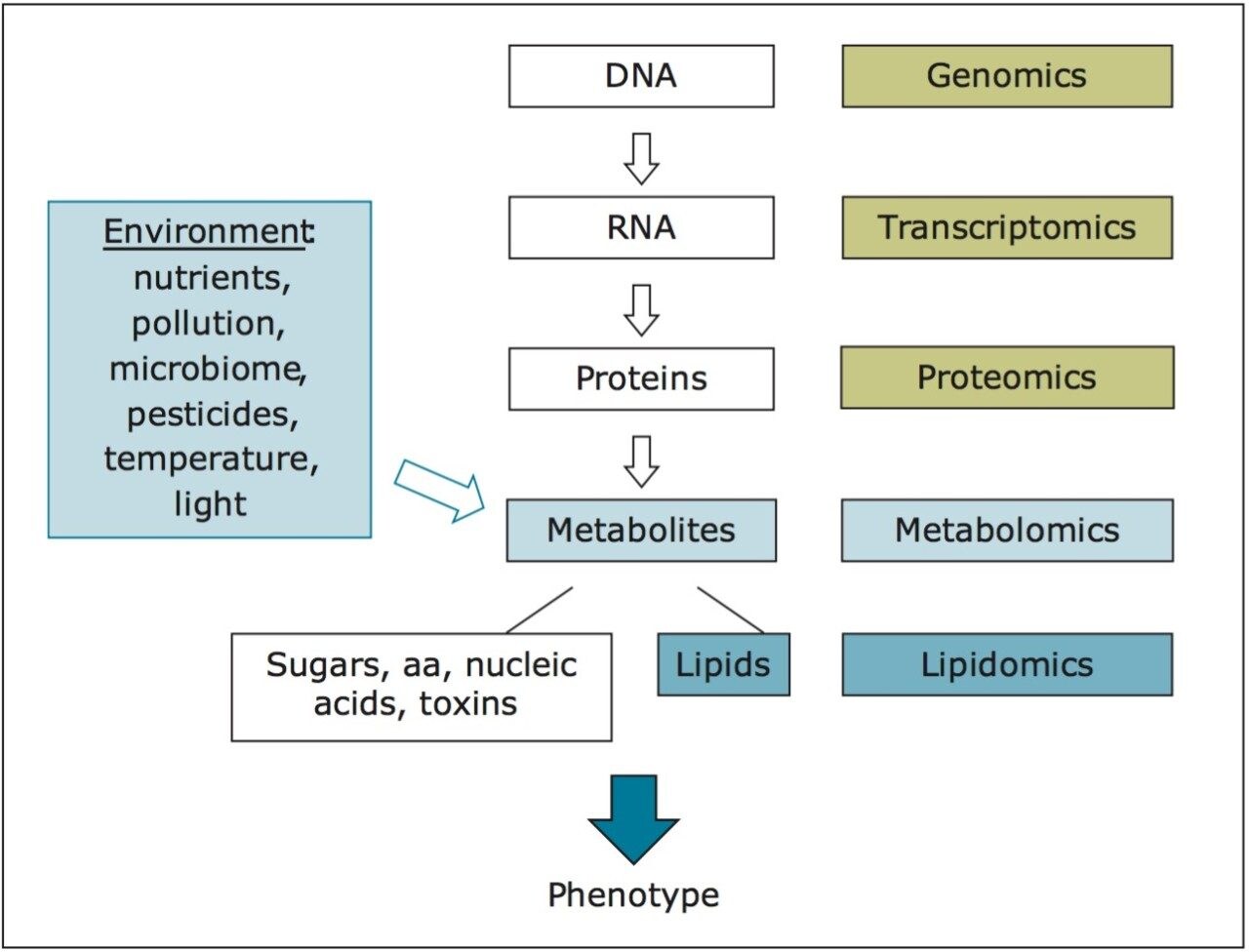
The aim of this work is to study how the complete set of small-molecule metabolites, the “metabolome,” of broccoli sprouts is modulated under different growth conditions. As the metabolome reflects both genetic and environmental components (e.g., light conditions and nutrients), comprehensive metabolite profiles can describe a biological system in sufficient depth to closely reflect the ultimate phenotypes, as shown in Figure 1.
Broccoli seeds (Brassica oleracea L. var. botrytis subvar. cymosa) were germinated in the germination cylinder of Vitaseed sprouter, and grown hydroponically for five days at 21 °C in a plant growth chamber (Clf Plant Climatics, Wertingen, Germany) equipped with PHILIPS Master TL-D 36W/840 cool-white fluorescent tubes providing a photosynthetic photon flux density of 110 mmol m-2 s-1, under three different light regimes: a) dark (achieved by covering the sprouting device with a cardboard box), b) continuous light, and c) continuous light plus two days of treatment with sucrose 176 mM.
Sprout samples, collected from the germination cylinder, were immediately frozen in liquid nitrogen and stored at -80 °C. Frozen sprouts were ground to a fine powder in a Waring blendor cooled with liquid nitrogen. Each sample of broccoli sprouts was extracted with 100% methanol (sample to solvent ratio 1:25 w/v) at 70 °C for 30 minutes under vortex mixing to facilitate the extraction. The samples were successively centrifuged (4000 rpm, 30 minutes, 4 °C), the supernatants were collected, and the solvent was completely removed using a rotary evaporator under vacuum at 40 °C. The dried samples were dissolved in methanol with the same volume of extraction, and filtered through 0.20-μm syringe PVDF filters.3
|
System: |
ACQUITY UPLC |
|
Column: |
ACQUITY CSH C18 2.1 x 100 mm, 1.7 μm |
|
Mobile phase A: |
60:40 10 mM NH4HCO2 in ACN/H2O |
|
Mobile phase B: |
90:10 10 mM NH4HCO2 in IPA/ACN |
|
Flow rate: |
0.4 mL/min |
|
Column temp.: |
55 °C |
|
Injection volume: |
5.0 μL |
|
Min |
A% |
B% |
Curve |
|---|---|---|---|
|
Initial |
60.0 |
40.0 |
Initial |
|
2.0 |
57.0 |
43.0 |
6 |
|
2.1 |
50.0 |
50.0 |
1 |
|
12.0 |
46.0 |
54.0 |
6 |
|
12.1 |
30.0 |
70.0 |
1 |
|
18.0 |
1.0 |
99.0 |
6 |
|
18.1 |
60.0 |
40.0 |
6 |
|
20.0 |
60.0 |
40.0 |
1 |
UPLC analytical column was connected to the ESI probe using PEEK Tubing, 1/16 inch, (1.6 mm) O.D. x 0.004 inch. (0.100 mm) I.D. x 5 ft (1.5 m) length, cut to 400 mm in length.
|
Mass spectrometer: |
SYNAPT G2-S HDMS |
|
Mode of operation: |
Tof HDMSE |
|
Ionization: |
ESI +ve and -ve |
|
Capillary voltage: |
2.0 KV (+ve) and 1.0 KV (-ve) |
|
Cone voltage: |
30.0 V |
|
Transfer CE: |
Ramp 20 to 50 V |
|
Source temp.: |
120 °C |
|
Desolvation temp.: |
550 °C |
|
Cone gas: |
50 L/h |
|
MS gas: |
Nitrogen |
|
IMS T-Wave velocity: |
900 m/s |
|
IMS T-Wave height: |
40 V |
|
Acquisition range: |
50 to 1200 |
TransOmics Informatics and HDMS Compare for SYNAPT Systems
We applied an untargeted metabolomics approach (Figure 1) to identify molecular alterations induced by different growth conditions in broccoli sprouts (Figure 2). Metabolites were extracted from the sprouts, and analyzed using UltraPerformance LC (UPLC) coupled with an ion mobility enabled QTof mass spectrometer, the SYNAPT G2-S HDMS (Figure 3), as reported in the Experimental conditions.
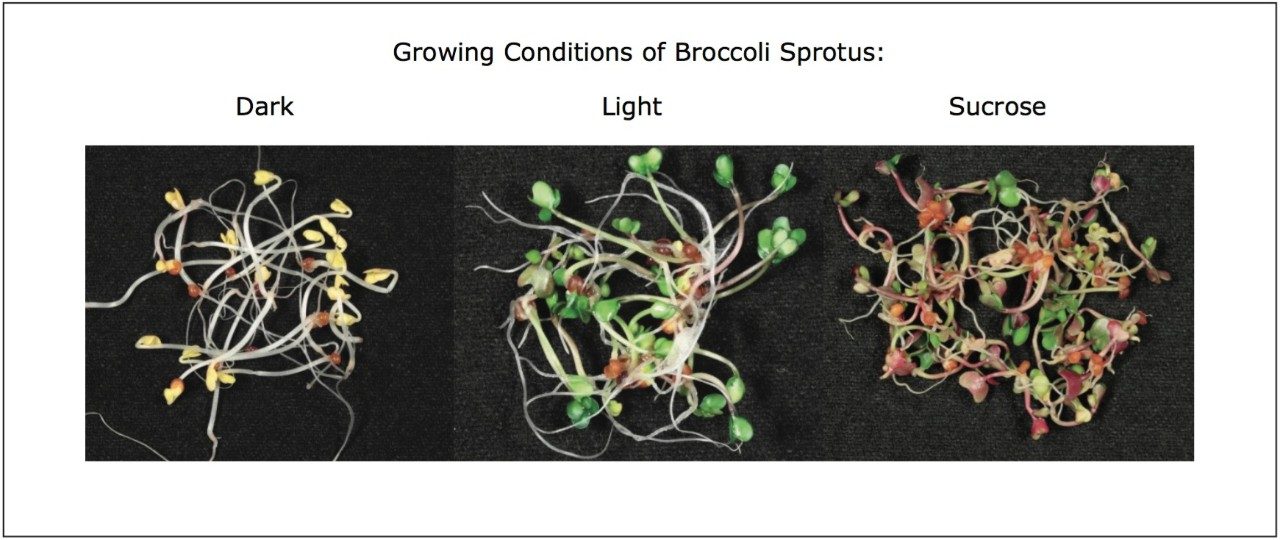
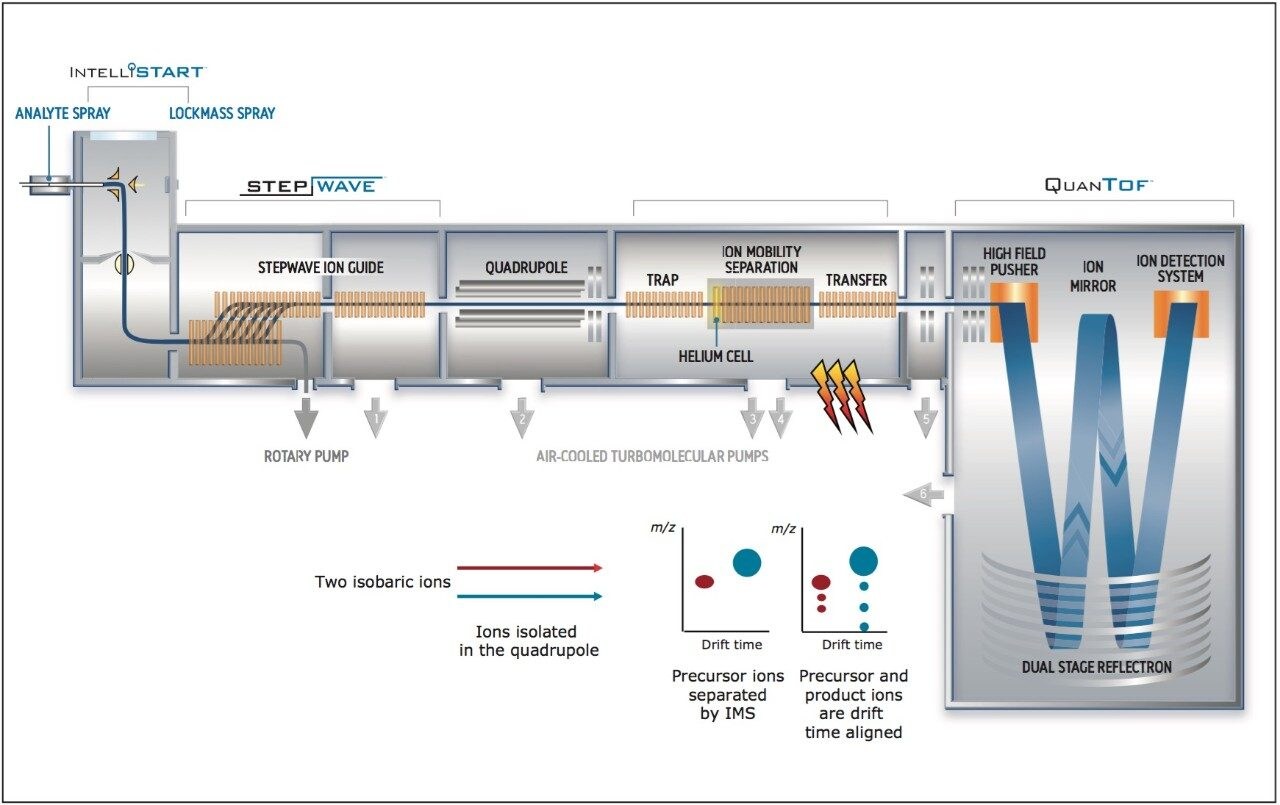
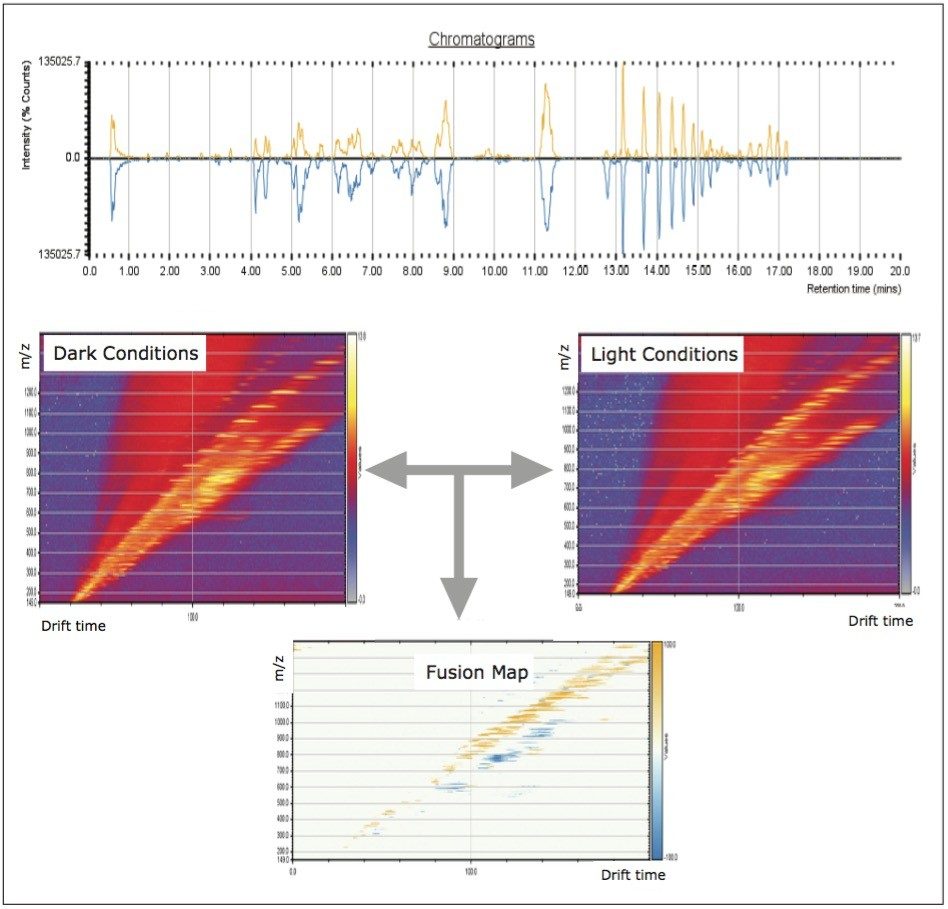
UPLC maximized the separation of a wide range of chemical complexity present in the broccoli sprouts (Figure 4). Metabolites were ionized using ESI and, subsequently entered into the vacuum region of the MS system where they passed through the tri-wave ion mobility separation (IMS) cell (Figures 3, 4, and 5).
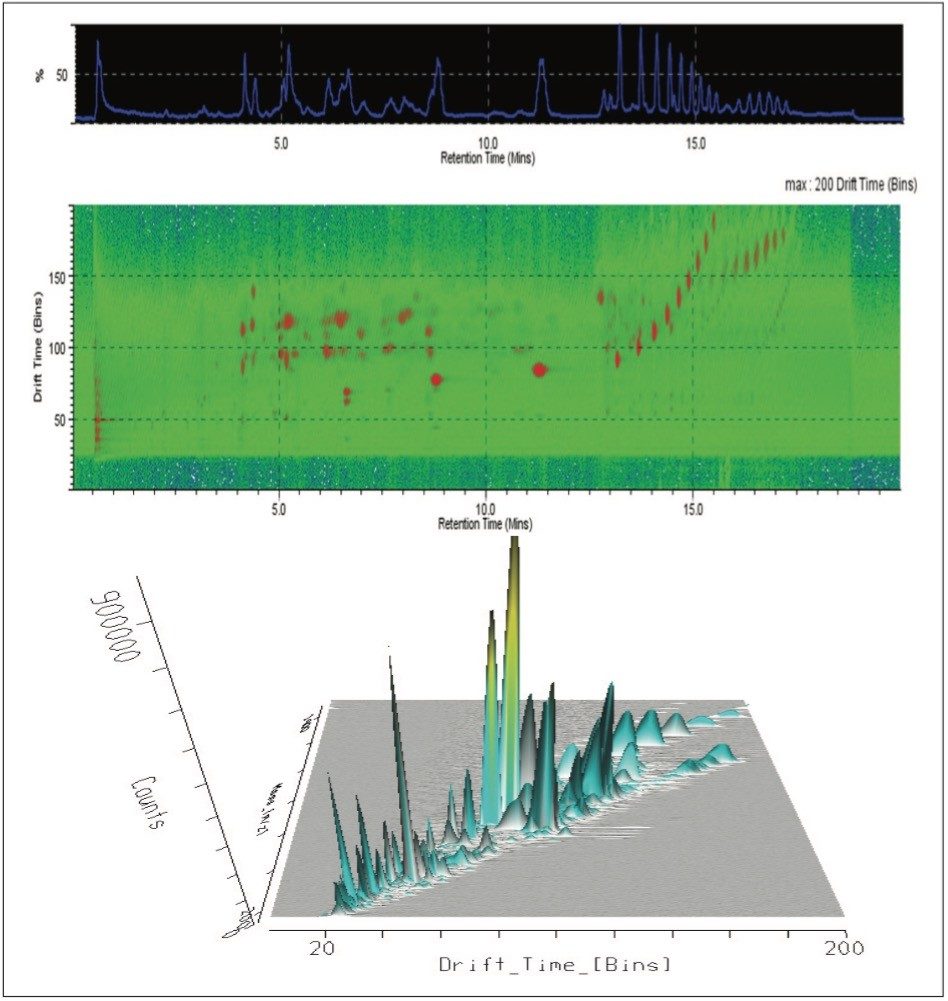
The T-Wave IMS device uses RF-confining fields to constrain the ions in the radial direction, while a superimposed repeating DC voltage wave propels ions in the axial direction through the dense gas-filled cell. The height and speed of the wave can be used to separate ions by their ion mobility1. As such, metabolites migrate with characteristic mobility times (drift times) according to their size and shape (Figures 3, 4, and 5). Therefore, IMS provides an additional degree of separation to chromatography, improving peak capacity over conventional UPLC-MS techniques (Figure 5).
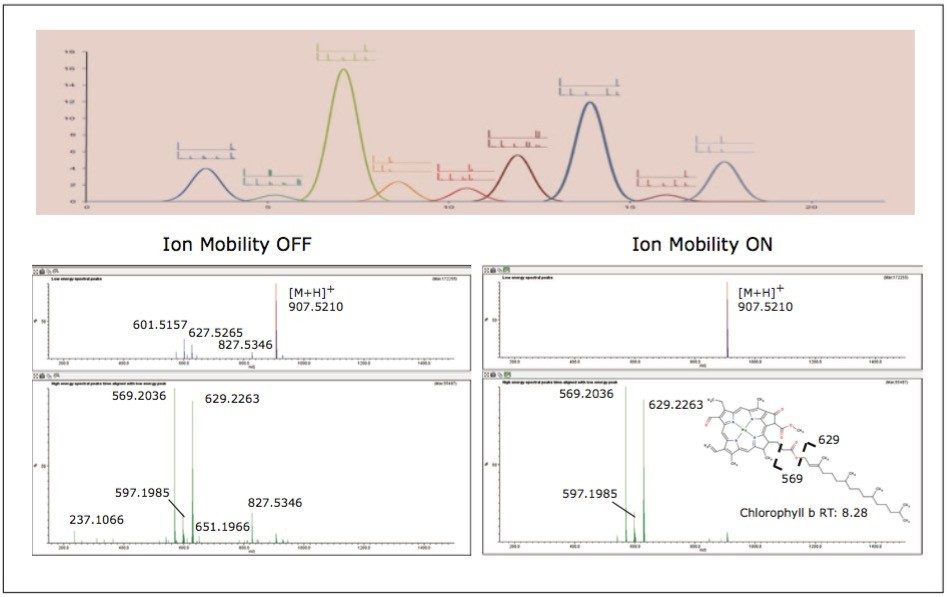
To aid in the identification and structural elucidation of metabolites, collision induced dissociation (CID) of metabolite precursor ions after IMS separation is performed using a particular mode of operation named HDMSE. This approach utilizes alternating low and elevated collision energy in the transfer cell, thus recording all of the precursor and fragment ions in a parallel and continuous manner (Figure 6). The alternating scans acquire low collision energy data, generating information about the intact precursor ions, and elevated collision energy data, that provides information about associated fragment ions (Figure 6). The incorporation of ion mobility separation of coeluting precursor ions before CID fragmentation produces a cleaner MS/MS product ion spectra, facilitating easier metabolite identification, as shown in the bottom panel of Figure 6.
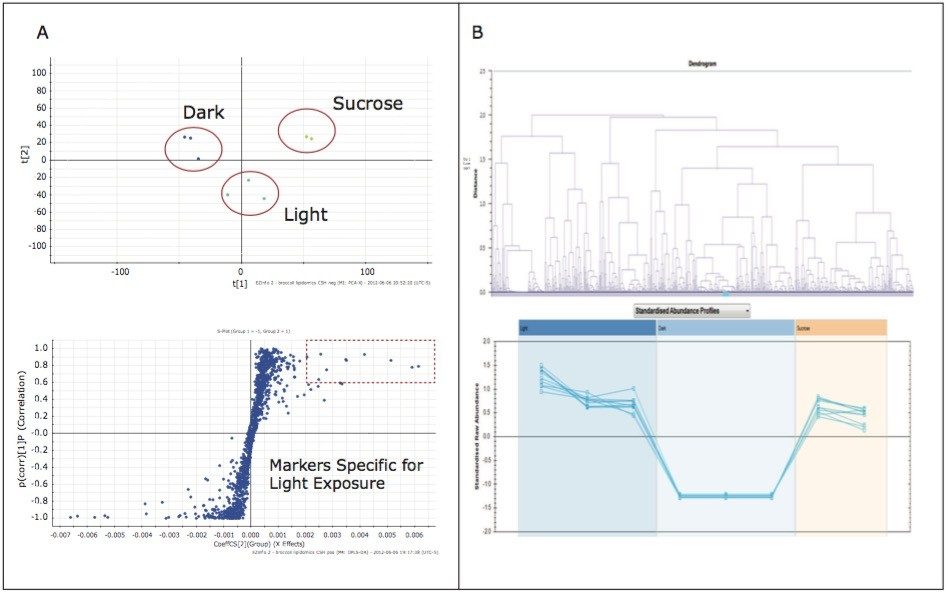
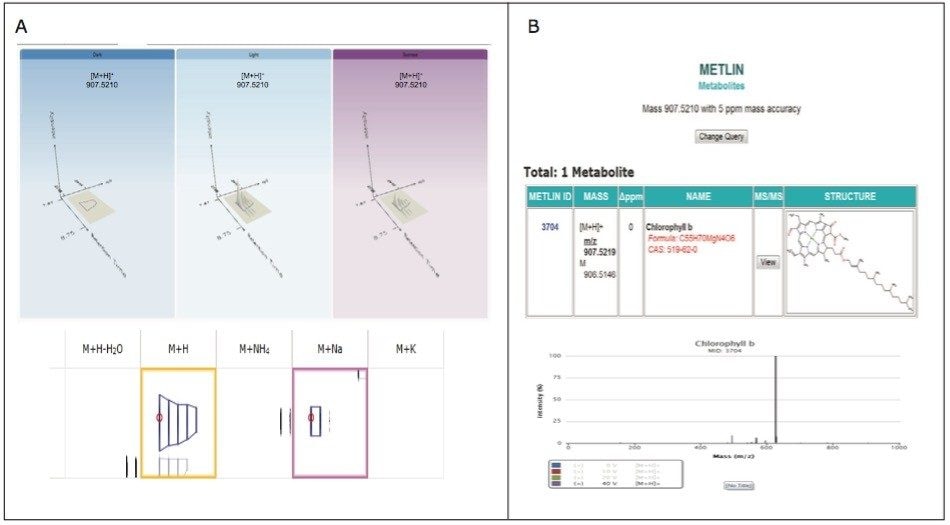
The analysis provided a metabolite profile, which represents a biochemical snapshot of the metabolite inventory for each sample analyzed. Differences at the metabolite level between groups were analyzed using TransOmics Informatics which provided multivariate statistical analyses tools, including principal component analysis (PCA) (Figure 7A), correlation analysis (Figure 7B), review compound (Figure 8A), and database search functionalities (Figure 8B) for metabolite identification.
Preliminary results suggest that growth conditions induce specific alterations in the “metabolome” of broccoli sprouts, some of which are strictly related to photosynthetic processes.
The Waters Omics Research Platform with TransOmics Informatics, featuring UPLC and HDMSE technologies, enables researchers to improve how they screen and differentiate molecular phenotypes of plants exposed to different environmental stimuli. This highthroughput approach has applications in agricultural, food, and nutritional, as well as natural product research.
720004703, June 2013