This application note demonstrates ETD-based MS technologies used for structural N- and O-glycan analysis, describing the modification site determination, and sequence information that can be readily obtained. A recombinant mAb, trastuzumab, is used as a test case for this study.
Protein glycosylation is a major requirement in the production of recombinant therapeutic antibodies and plays a vital role in its efficacy and safety. Typical glycosylation analysis consists of a released glycan assay, glycosylation site determination, and site heterogeneity analysis.
Throughout the years, biopharmaceutical laboratories have developed and optimized the glycan profiling analysis using a variety of liquid chromatographic separation modes with optical detectors. Tandem mass spectrometry based approaches represent a rapid and sensitive approach to obtain site-specific characterization of glycosylation. Electron transfer dissociation (ETD) is a powerful fragmentation technique known to be particularly useful for determining modification sites of labile post-translational modifications (PTMs), which are often more difficult to characterize using collision induced dissociation (CID). This application note offers details of glycopeptide characterization using ETD fragmentation.
Selected precursor ion charge-state is important in ETD, since this affects the dissociation efficiency.1 Non-dissociative electron transfer is often observed as a function of decreasing precursor ion charge; therefore, m-nitrobenzyl alcohol (m-NBA) was added post-column to increase the charge-state of the ESI-generated ions to enhance ETD efficacy.2
We demonstrate ETD-based MS technologies used for structural N- and O-glycan analysis, describing the modification site determination, and sequence information that can be readily obtained. A recombinant mAb, trastuzumab, is used as a test case for this study.
|
Mass Spectrometer: |
SYNAPT G2-S HDMS |
|
Capillary: |
2.0 kV |
|
Reagent glow discharge current: |
70 μA |
|
Source temp.: |
100 °C |
|
Sampling cone: |
25 V |
|
Extraction cone: |
3 V |
|
Reagent make-up gas flow: |
25 mL/min |
|
Desolvation temp.: |
200 °C |
|
Cone gas flow: |
30.0 L/h |
|
ETD reagent: |
1,3-dicyanobenzene (m/z 128) |
|
Trap wave height: |
0.25 V |
|
Trap RF: |
500 V |
3-Nitrobenzyl alcohol (m-NBA), was purchased from Fluka (Part number 73148), and the ETD reagent, 1,3-Dicyanobenzene, was purchased from Sigma-Aldrich (Part number 145858). Trastuzumab was reduced and alkylated using diothiothreitol and iodoacetamide, followed by overnight trypsin digestion.
An ACQUITY UPLC H-Class Bio System was directly coupled to the standard ESI interface of a SYNAPT G2-S HDMS Mass Spectrometer equipped with ETD. Reversed phase chromatographic separation of 1 to 10 pmol of tryptic peptides was performed on an ACQUITY UPLC BEH300 C18, 2.1 x 150 mm, 1.7-µm Column. Mobile phases A and B were water and acetonitrile, both containing 0.1% formic acid. The tryptic peptides were eluted from the column using a gradient from 5% to 35% B over 35 min at a flow rate of 100 µL/min. The gradient was further ramped to 50% B for 10 min, then to 80% B for 1 min, and held for 5 min at a flow rate of 200 µL/min. Post-column addition of ~0.4% m-NBA, a ‘super-charging’ reagent, was achieved using a Valco T-piece.
ETD has been used as a complementary tool to CID for protein post-translational modification analysis. CID fragmentation of glycopeptides is known to favor neutral losses (cleavage of regions within the attached sugar); consequently, intense oxonium ions are detected at the low m/z. Since CID is usually dominated by glycan-related product ions, this hampers peptide sequence information for precise site-specific identification of this important class of PTM.
Fortunately, ETD fragmentation follows a different fragmentation mechanism where the glycan moieties are preserved on the c and z fragment ions. Since fragmentation is conducted post separation, these fragment ions are generated at the same retention time as those in the glycan moiety CID analysis.
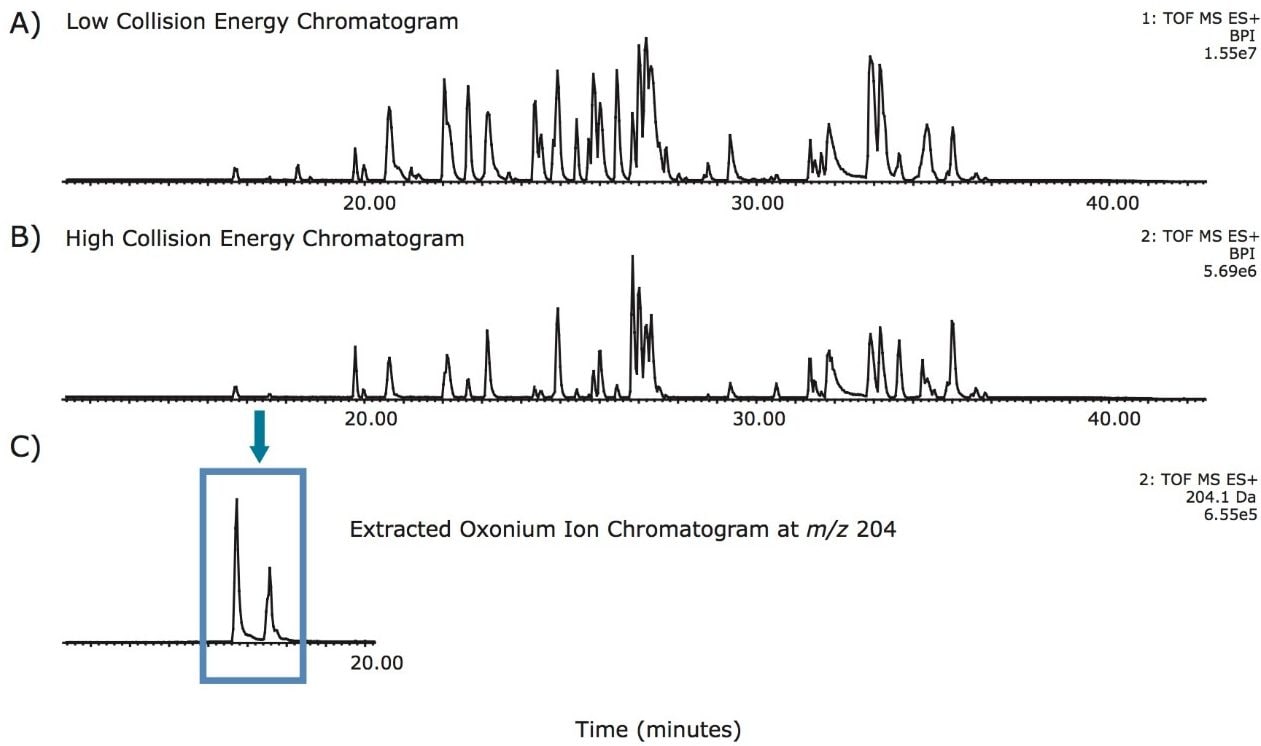
Tryptic recombinant monoclonal IgG, trastuzumab, was used for the ETD analysis. Trastuzumab is known to have one N-linked glycosylation site on the heavy chain in the Fc region. Glycopeptide chromatographic retention times were determined from a UPLC-MSE experiment. Since the common oxonium ions, e.g., m/z 204.09, 366.14, from the high energy fragmentation data channel are the dominant fragments, as shown in Figure 1, the m/z extracted ion chromatograms allow identification of the retention times of the glycopeptides and obtain their m/z values. The retention times and precursor m/z values were used to set up the subsequent ETD experiment.
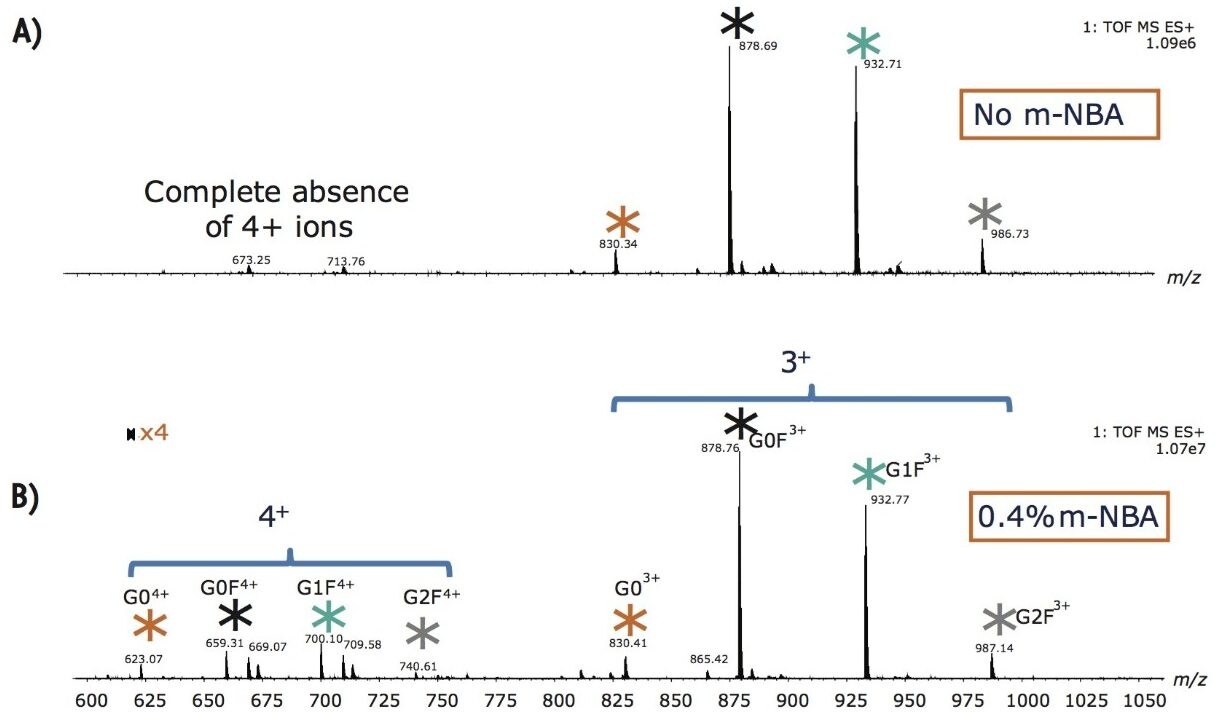
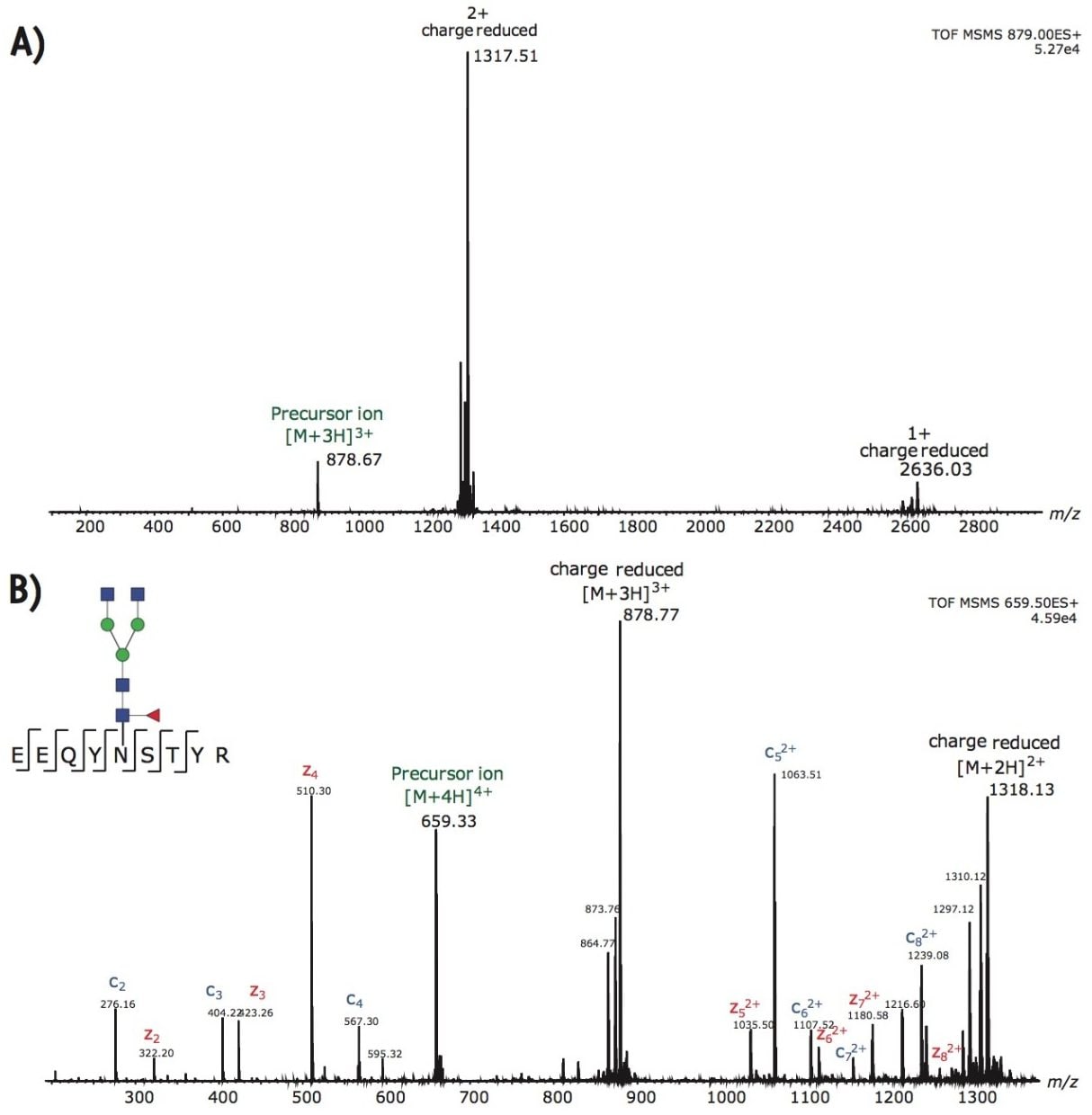
By post column mixing the super-charge reagent, m-NBA, the overall charge envelopes of the targeted glycopeptides are shifted to favor higher charges, as shown in Figure 2. Non-dissociative electron transfer is often observed as a function of decreasing precursor ion charge. Figure 3A shows the fragmentation of the triply charged precursor ion (peptide contains G0F) which generates charge-reduced to 2+ and 1+ ions, with little peptide backbone fragmentation. Figure 3B demonstrates that intense c and z ions were produced when m-NBA was used to promote the quadruply charged precursor ion for ETD. The net mass difference between the c4 to c5 (and z4 to z5) ions matches the mass of Asn plus G0F (total mass difference of 1558.6 Da).
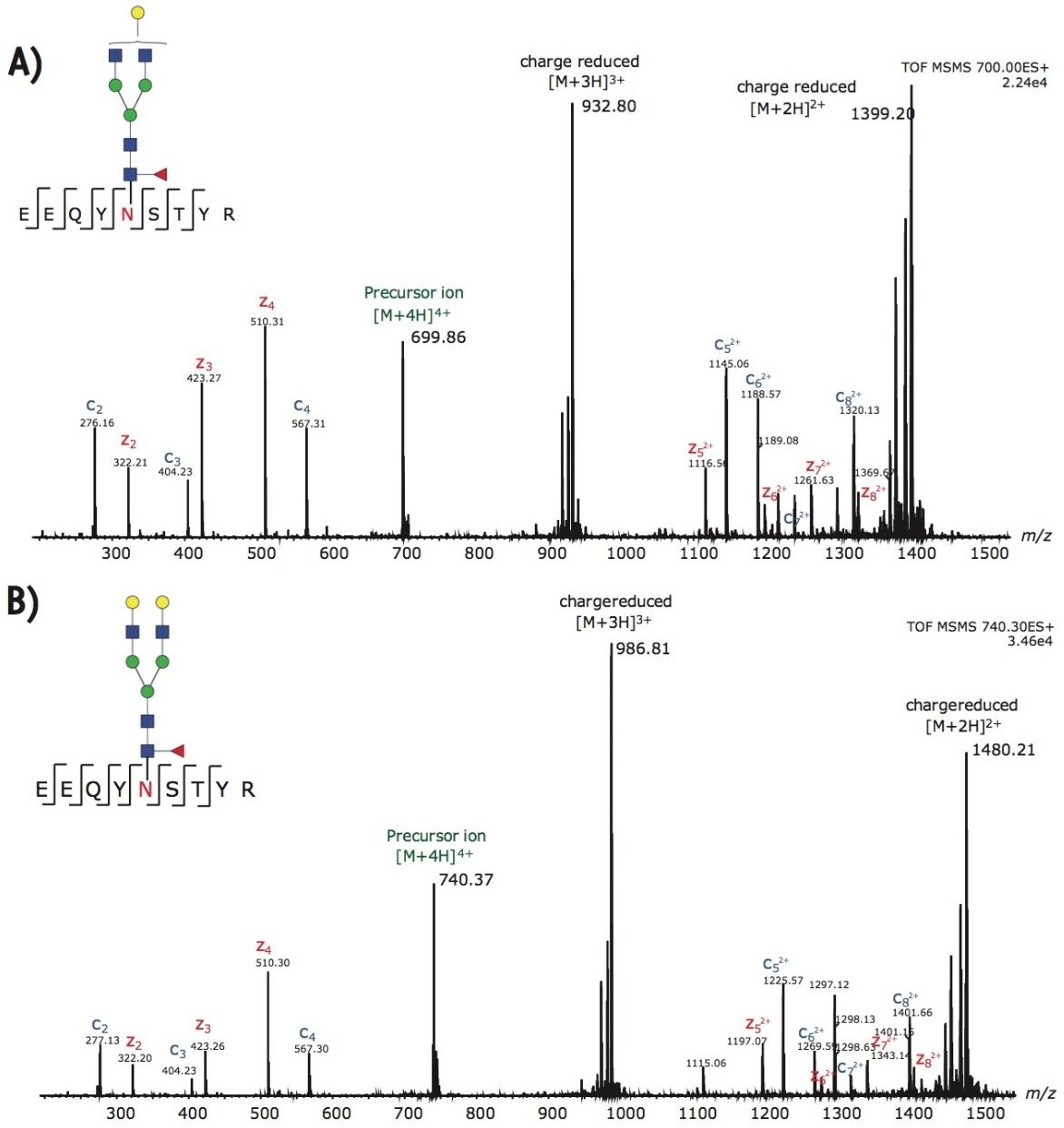
In addition to G0F, the other two dominant glycoforms from trastuzumab are G1F and G2F. Figures 4A-B illustrate the ETD fragmentation of the quadruply charged precursor ions from these glycopeptides. The c and z ions in these spectra are used to confirm the peptide backbone with high confidence. In addition, we observed little to no neutral loss from the glycan moiety; hence, the intact glycan mass can be assigned to the glycosylation site with single amino acid resolution.
ETD proved to be an indispensable tool for characterizing post translational modifications of biotherapeutic proteins. The data from this application demonstrate that glycopeptide site heterogeneity from a monoclonal antibody, trastuzamab, can be achieved using ETD on a SYNAPT G2-S HDMS System with post-column mixing of m-NBA.
720004481, October 2012