For research use only. Not for use in diagnostic procedures.
This is an Application Brief and does not contain a detailed Experimental section.
This application brief demonstrates an automated and universal UPLC-HDMSE workflow for the identification and characterization of the major classes of lipid molecular species from a complex biological sample.
UPLC separations, ion mobility and advanced informatics combine to give unique insight into your lipid profiling studies.
Lipids are both the building blocks and main repository of energy in cell membranes. Recent studies have shown that lipids can also play an essential role as signaling molecules and have the potential to revolutionize biomarker discovery and future diagnostic testing for various diseases. Mass spectrometric-based global lipid profiling techniques are diverse in both method of sample introduction and detection. Direct infusion methods, or those based upon normal phase chromatography have prevailed historically due to the apparent simplicity and applicability of the method. Similar considerations have also led to nominal mass instruments being the preferred detection platform for lipid interrogation. However, when choosing any analysis technique it is important to consider the complexity of the biological sample. This means factoring in issues, such as ion suppression from high abundant lipid species and the need for detection and quantification of low abundance and isobaric lipid species.
In this technology brief we will present a novel platform based upon reverse phase UPLC and exact mass MS detection that aims to address these challenges with a simple, comprehensive, and automated novel UPLC-TOF HDMSE lipid discovery workflow using SYNAPT G2 and SYNAPT G2-S systems. We will demonstrate the workflow using bovine liver total lipid extract to maximize the amount of information obtained using UPLC-HDMSE analysis.
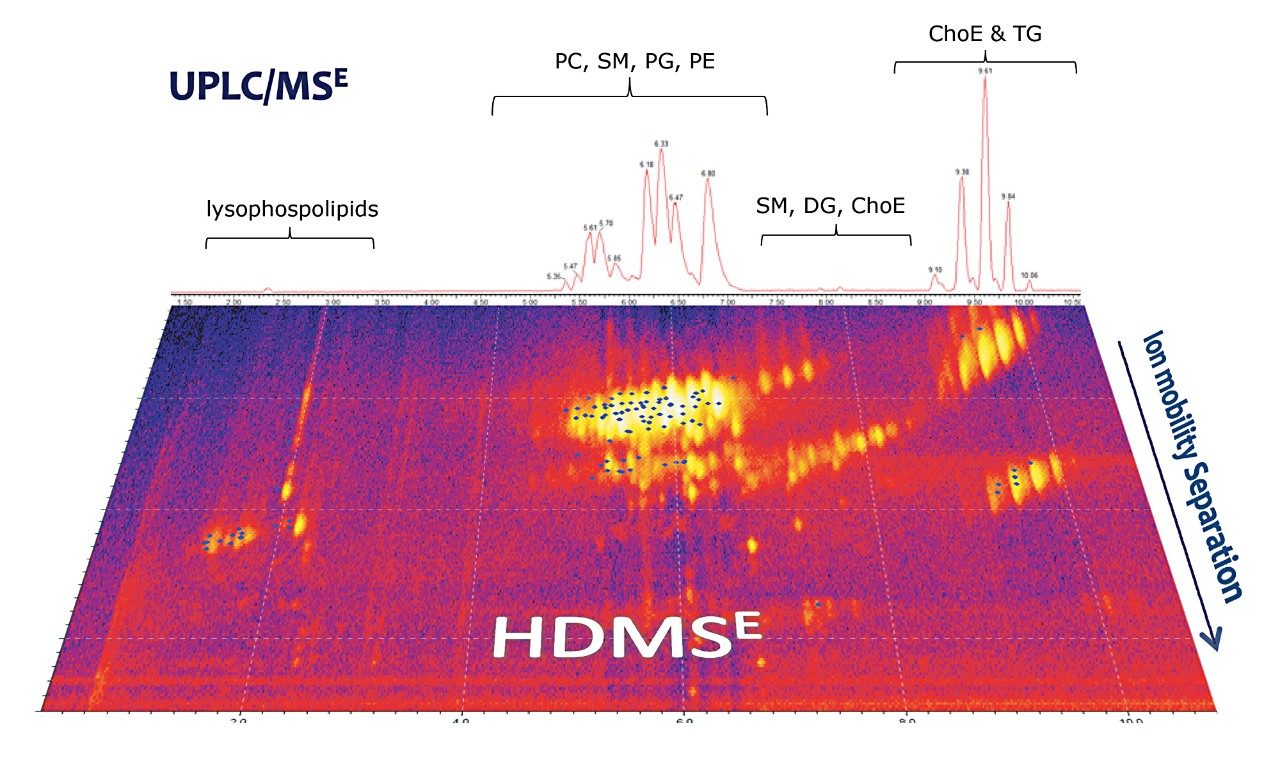
For this study, total liver extract was used in order to highlight the extraction of data of exceptional quality from a complex matrix using straightforward, robust methodology. A working 0.1 mg/mL solution of liver extract was prepared by diluting the stock solution (5 mg/mL in chloroform/methanol, 2/1) with isopropanol/acetonitrile/water (50/25/25). From a single analysis, comprehensive datasets were collected using UPLC-HDMSE with a SYNAPT G2 System. The UPLC method used for this lipid separation is based on ACQUITY UPLC HSS T3 Column Technology. The highly efficient UPLC separations provided excellent reproducible performance for lipid analysis in a short 15-minute timeframe, as shown in Figure 2. This separation was further enhanced with Ion Mobility Separation (IMS), which provides an added dimension to the analysis by separating isomeric lipids based on their size and shape.
The HDMSE acquisition method utilizes parallel low and elevated collision energy which provided precursor and product ion information for every detectable component in the mixture. In order to utilize the HDMSE data, Apex 4D, a novel automatic peak peaking algorithm, was employed to extract and align the low and elevated data by m/z, retention time, and drift time. MSE Data Viewer Software then allows a user to visualize, manipulate, and export the data to SimLipid for further lipid identification, as shown in Figure 3.
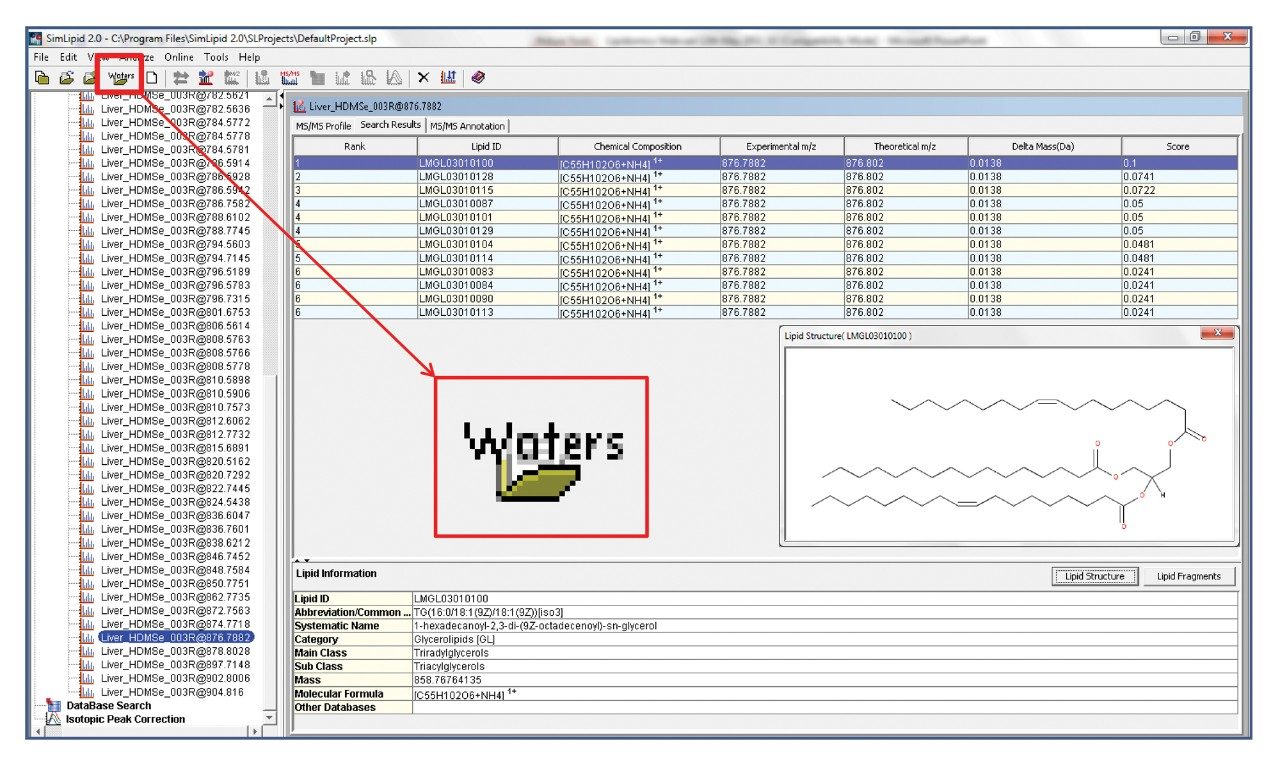
SimLipid contains a database of over 22,000 lipids and completes the workflow, as shown in Figure 1, by automatically searching the database using precursor and product ion information gathered from the HDMSE dataset. The returned database hit is scored based upon the match of observed to in-silico predicted fragments. The results can then be reported and exported in a variety of ways, including .csv, .xls, and HTML outputs.
The workflow detailed in this application brief provides an automated separation, identification, and characterization solution for global lipid analysis without any prior knowledge of the sample and has the potential to speed the rate at which unknown lipids are identified.
Combining MSE and IMS separation (HDMSE) with conventional reverse phase chromatography allows scientists to analyze difficult lipid samples with complex isobaric/isomeric properties.
For the first time, this UPLC-HDMSE solution provides scientists with an easy-to-use and highly specific lipid analysis method that gives robust and reproducible results.
720004000, May 2011