This is an Application Brief and does not contain a detailed Experimental section.
This application brief demonstrates the reproducibility and applicability of the Waters ACQUITY UPLC System with Xevo G2-XS QTof MS for the analysis of biological fluid resulting from a large cohort metabolic profiling study.
The combination of the ACQUITY UPLC System and Xevo G2-XS QTof Mass Spectrometer demonstrates excellent reproducibility of peak retention and peak area – making it suitable for successful metabolic profiling of human urine in large cohort biomedical research.
The generation of biologically-meaningful information in biomedical research or epidemiological studies requires large cohort studies to overcome any skewing of the data due to subject age, gender, diet, etc. Equally as important is the utilization of an analytical technique that ensures the sample analysis exhibits as minimal a variation or drift as possible. From a statistical point of view, the lower the analytical variation, the greater the likelihood of subtle biological variation being observed. In an LC-MS analysis, analytical variation typically manifests as retention time drift, mass accuracy drift, or peak area fluctuation. The presence of any variation may result in irreproducibility, QC failure, or obscuring of subtle, biologically-significant features.
To evaluate the suitability of the Waters ACQUITY UPLC System with Xevo G2-XS QTof MS for the analysis of biological fluids from a large-scale cohort study, the system was challenged with the analysis of greater than 1300 injections of human urine. A bulk analytical sample was prepared and two stable labeled internal standards were spiked in the sample to act as reference markers. A QC sample was prepared using the same control bulk urine sample as with the analytical sample, with an additional eight stable labeled internal standards spiked into the QC mix.
The samples were aliquotted into 96-well plates with sufficient analytical sample for triplicate injections from each sample well, and sufficient QC for a single injection per QC well. Two sample wells are followed by one QC well through the plate, giving a total of 224 injections per plate with a QC analysis every seventh injection.
The analysis conducted was reversed-phase chromatography employing a 1.8 µm, 2.1 mm x 150 mm ACQUITY UPLC HSS T3 Column. The injections were eluted with an aqueous formic acid–acetonitrile gradient from 1 to 55% over ten minutes at a flow rate of 600 µL/min. The gradient elution was followed by a high-organic wash and re-equilibration. The column effluent was monitored by a Xevo G2-XS QTof Mass Spectrometer operating in positive ion electrospray mode. The mass spectrometer was operated in sensitivity mode with the synthetic peptide leucine enkephalin employed as the lock mass reference solution.
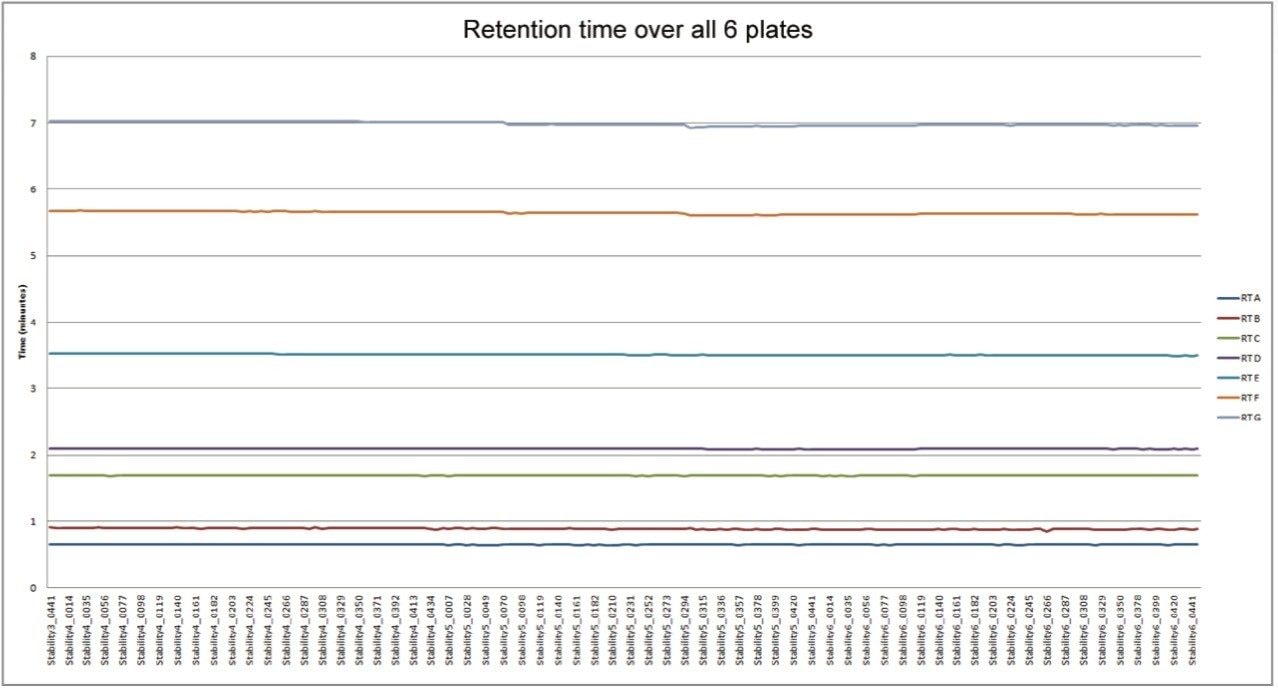
The resulting data for chromatographic retention time reproducibility is displayed in Figure 1. The data showed a relative standard deviation between 0.98% and 0.15% for the seven compounds measured in the QC injections from six 96-well microtiter plates. This data provides great confidence that the chromatography system delivered more than sufficient accuracy and precision for the task of metabolic phenotyping across a large sample batch.
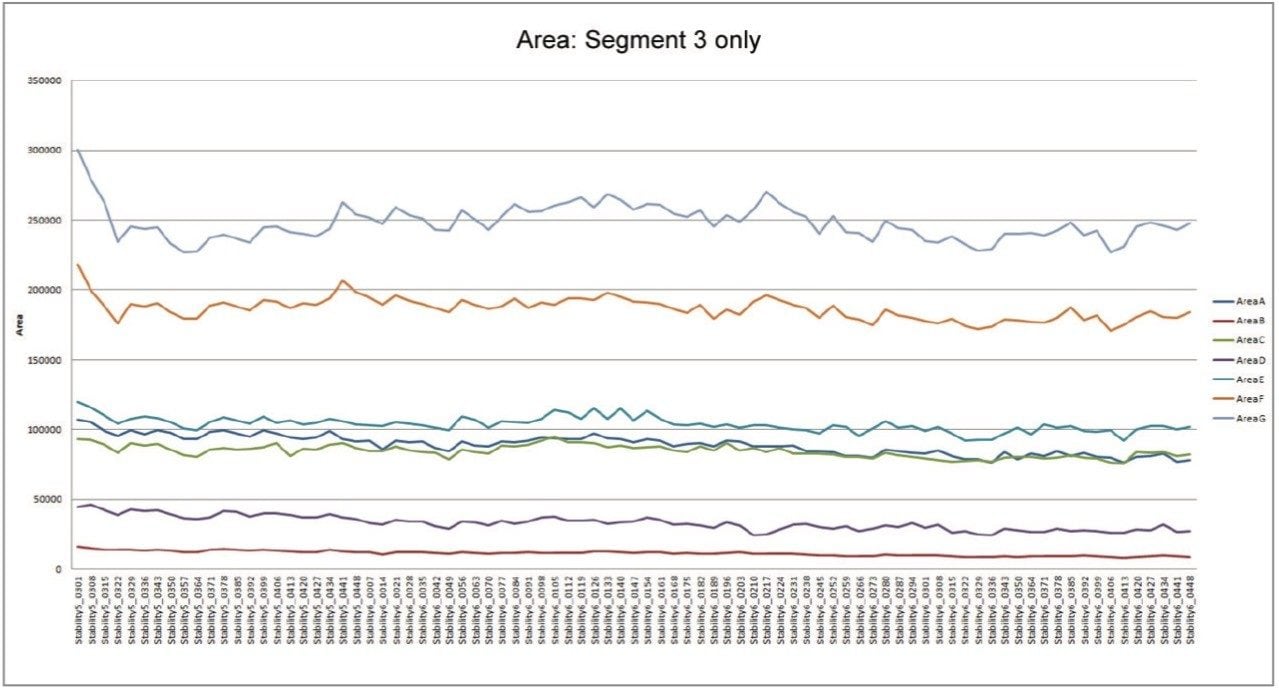
The statistical analysis tools employed in metabolic profiling – such as principal components analysis, partial least squares discriminant analysis, and orthogonal partial least squares discriminant analysis – rely on accurate assignment of component identity and assessment of the concentration of each component in the derived data. The variation in the mass spectrometry response over the duration of the analytical process for the seven compounds is shown in Figure 2. The coefficient of variation in response for the triplicate analysis of the QCs across three microtiter plates ranged from 4.2% to 15.5% with a mean variation of 8.3%.
This study demonstrated the reproducibility of the ACQUITY UPLC System with the Xevo G2-XS QTof Mass Spectrometer and its suitability for metabolic phenotyping. The system showed excellent retention time reproducibility with the standard variation being less than 1% for all compounds processed across greater than 1300 injections. The mass spectrometric peak area response showed a mean variation of 8.3% for the same compounds. This data demonstrates that the system is ideally suited to the analysis of large cohort studies of human biological fluids.
720005798, November 2016