This Application brief demonstrates wide dynamic range capabilities with improved spectral clarity when using SONAR to increase identification and quantitative accuracy of low level components in complex samples.
SONAR demonstrates high quantitative accuracy over a wide dynamic range for highly complex samples comprising of spiked components at low levels.
Quantitative lipidomics for various biological samples such as plasma and tissue extracts are routinely performed using LC-MS methodologies, with the most common form using multiple reaction monitoring (MRM) for targeted analyses. However, advancements in high resolution technology and the introduction of alternative data acquisition modes such as data independent acquisition (DIA) provide additional benefits over more traditional modes of acquisition. The complexity of lipid extracts provides technical challenges such as co-elution of multiple species, which consequently has an impact on false positive identification and quantitative accuracy. The presence of multiple fragment ions generated from the dissociation of co-eluting precursors can result in false estimates being reported for quantitative studies. SONAR is a DIA acquisition method that collects both precursor and MS-MS data, providing clarity without the associated method development steps. Here we describe the use of SONAR for targeted based studies, involving plasma lipid extracts spiked with deuteruated standards at varying concentration to assess the limits of quantitation, linearity and dynamic range.
SONAR has been utilized for the analysis of lipid extracts derived from pooled human plasma samples. Lipids were extracted using IPA (1:2 v:v) and incubated at room temperature for 10 min prior to overnight storage at -20 °C. Samples were then centrifuged at 14,000 g for 20 min and the resulting supernatant used for analysis. For quantitative purposes, deuterated lipid standards (SPLASH LipidoMix, Avanti Polar Lipids, Inc.) containing a range of lipid classes from 350 to 2 μg/mL were spiked into the plasma extracts at ratios ranging from 1:1 to 1:100,000 (Figure 1).
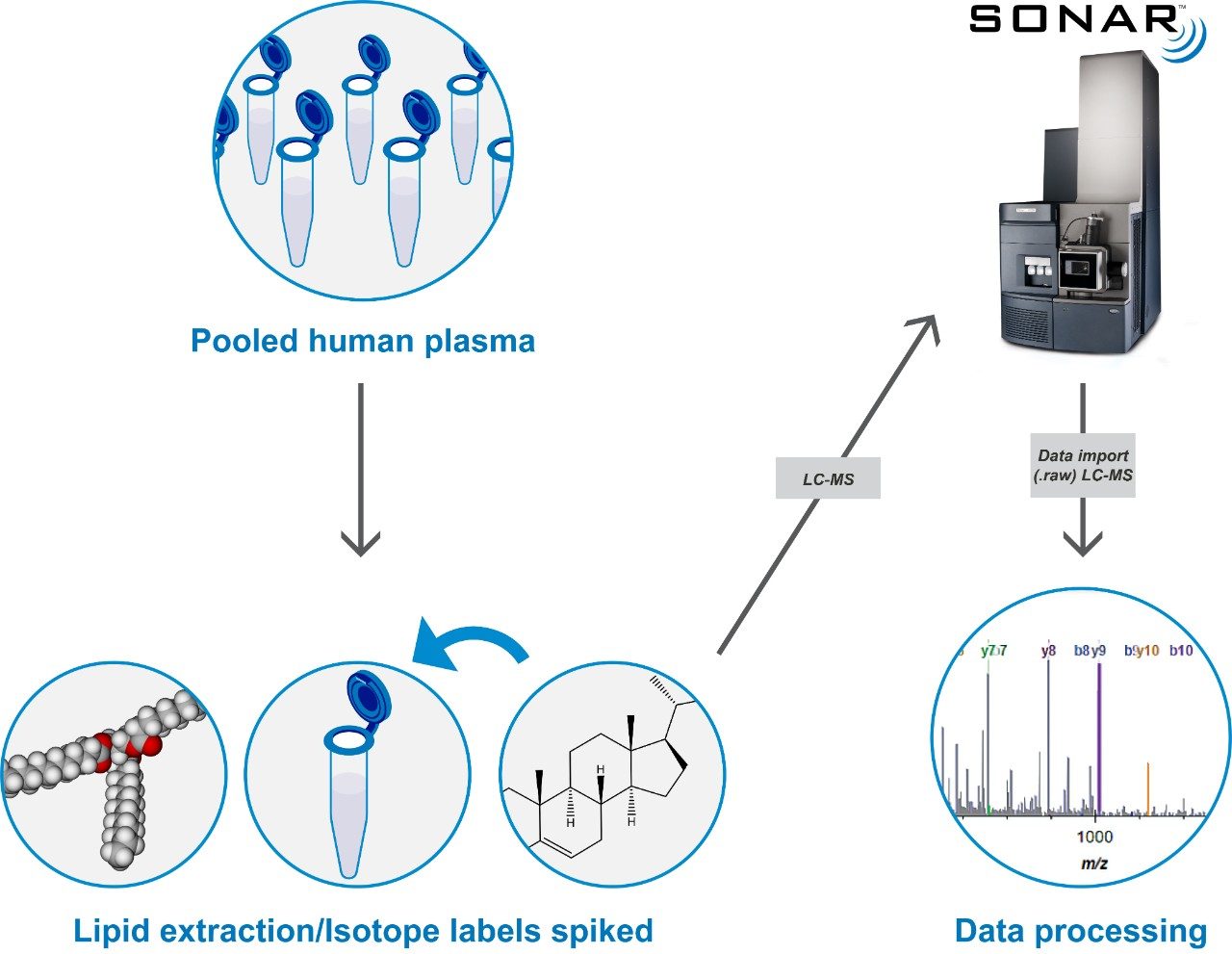
Lipids were separated over a 20 min LC gradient using reversed phase (RP) chromatography (ACQUITY UPLC CSH C18 Column, 1.7 μm, 2.1 x 100 mm).1 LC-MS data were acquired using a Xevo G2-XS QTof Mass Spectrometer operating in SONAR mode with a quadrupole window of 10 Da, scanning over a precursor mass range of 350–950 m/z, whilst the Tof scanned over 50–1200 m/z with a scan rate of 0.1 sec. Comparative DIA datasets based on the same sample loadings were also acquired using MSE, which operates a non-resolving quadrupole. Data from all experiments were processed and interrogated using Skyline (University of Washington) as shown in Figure 2.
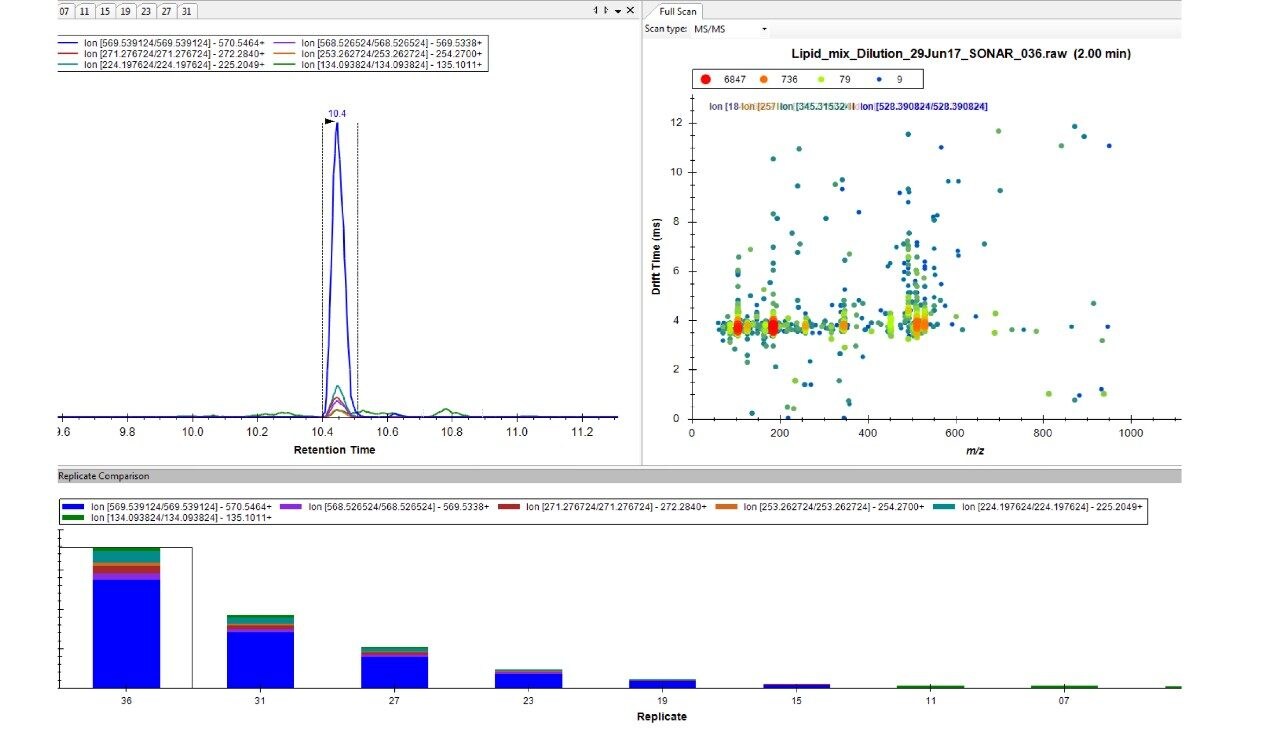
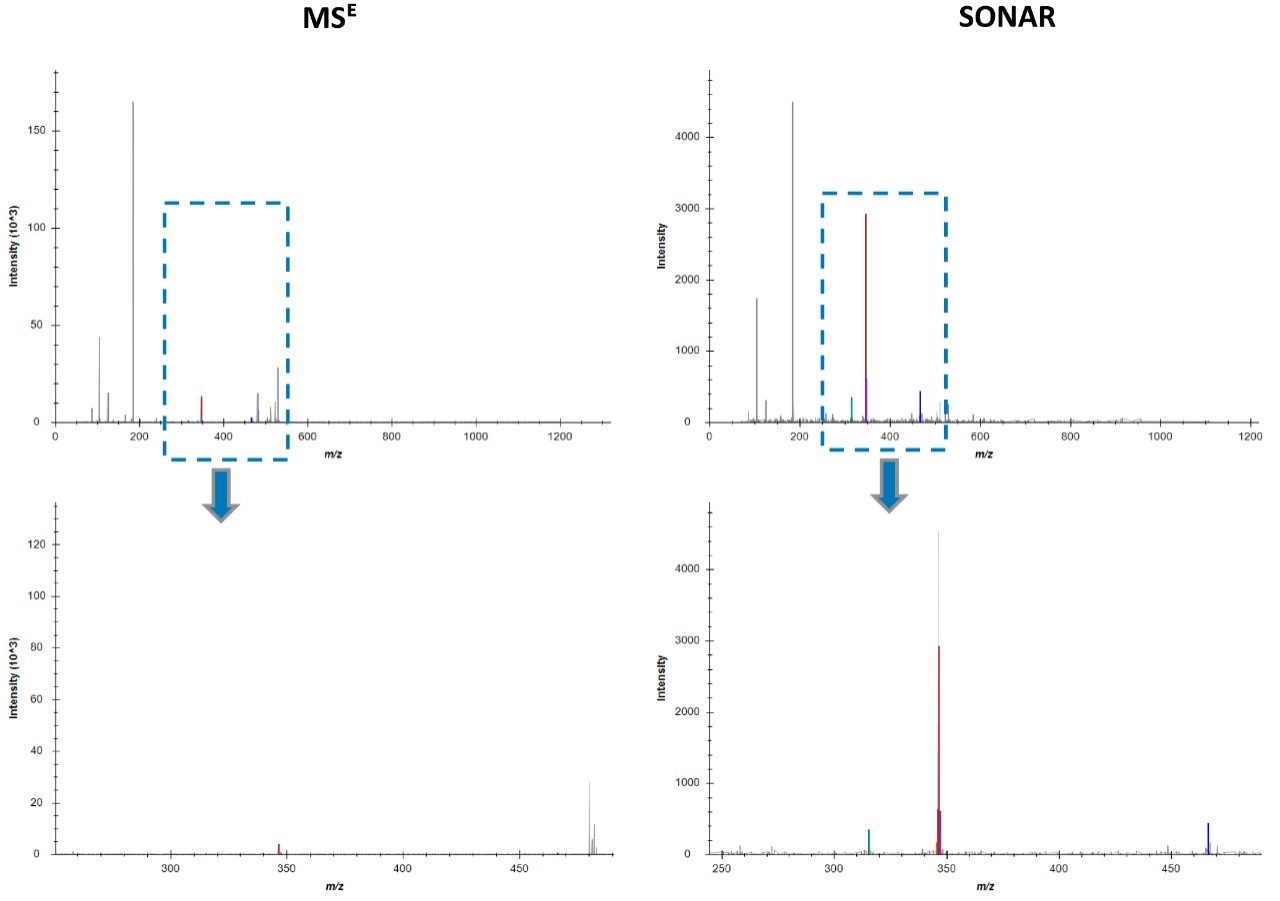
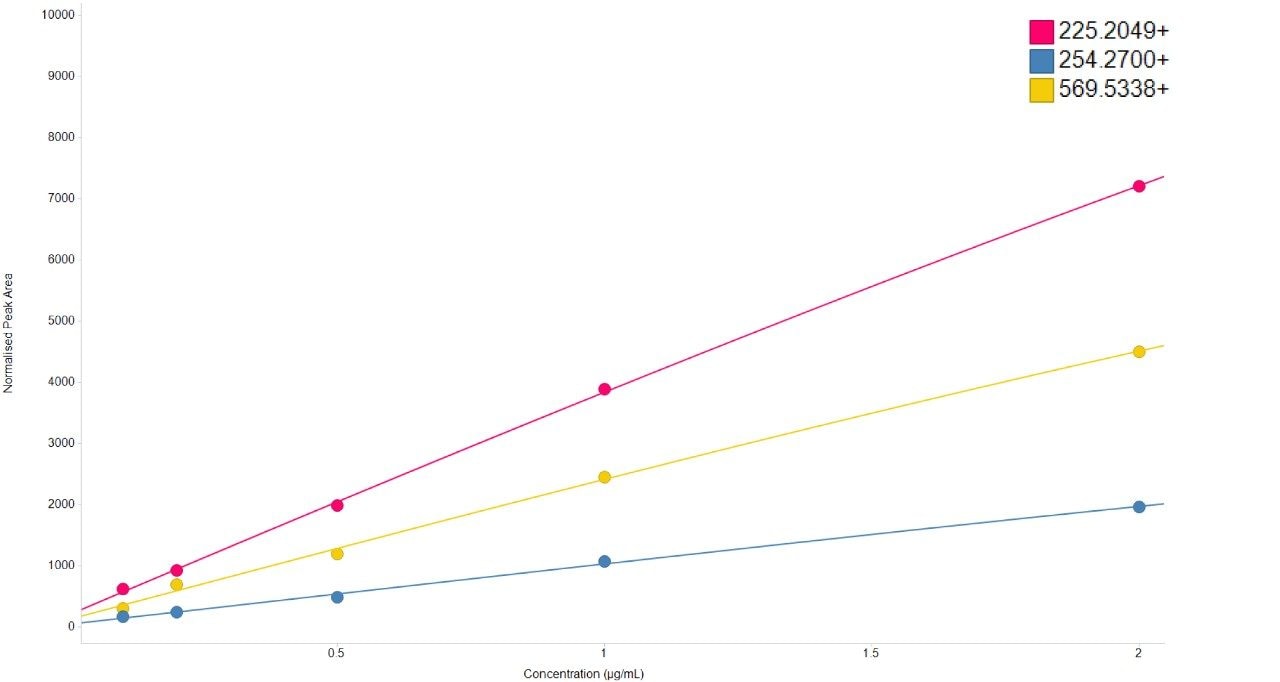
Chromatographic separation of complex lipid samples can result in the co-elution of multiple species, providing multiple precursors and therefore generate mixed fragmentation spectra. Inherently this impacts on the accuracy of quantitation which can be achieved due to the interference effects incurred from multiple fragment ions being present. An example of the spectral clean-up provided by SONAR is shown in Figure 3 for a representative LPE (18:1) (d7). On average, the limit of detection achieved (LOD) was 0.001 μg/mL, while 0.1 μg/mL was established as the limit of quantification (LOQ), which is comparable with previous targeted studies.2 The consequences of improved spectral clarity1 and quantitative accuracy are further demonstrated in Figure 4 with three transitions corresponding to DG (15:0/18:1) (d7) showing good linearity over a wide dynamic range.
This study demonstrates the wide dynamic range capabilities of SONAR with the detection of low level components (LOD=0.001 μg/mL) at high specificity. Interrogation of SONAR data through third party informatics tools such as Skyline, demonstrate a flexible workflow which can be applied to targeted approaches. SONAR is shown to generate qualitative and quantitative data with high selectivity whilst maintaining high duty cycle to in order to provide high quantitative accuracy. Data is collected as full scan DIA therefore providing flexibility for data analysis (i.e., the option to quantify using a wide selection of transitions). The concept of SONAR DIA ultimately allows for high throughput analysis and demonstrates its applicability for large cohort studies.
720006296, April 2018