
In this study, nefazodone and its metabolites were used as a model system to explore the advantages of IMS-enabled DDA using Vion IMS QTof Mass Spectrometer.
Co-eluting metabolites attributed to glucuronides of dihydroxylated metabolites were successfully characterised using IMS-enabled DDA, generating two distinct precursor ion MS spectra and product ion MS/MS spectra for the drift time separated metabolites. The m/z and drift time filtered data provide cleaner, unambiguous spectra and increases confidence in structural assignment compared with simple m/z-selective DDA.
LC-MS/MS using data-dependent acquisition (DDA) is widely used for characterization of metabolites in drug metabolism studies. In a DDA experiment, a full scan MS experiment is evaluated for trigger properties which then switch the instrument from MS to MS/MS. In the conventional DDA experiment, these triggers are typically m/z, charge state, isotope pattern, and retention time. In the situation where singly charged, isobaric, and near isobaric metabolites co-elute, the resulting MS/MS spectra will contain product ions from all precursors, presenting a challenge for interpretation.
The incorporation of ion mobility separation (IMS) measurements into DDA experiments, allows the product ion spectra of co-eluting isobaric metabolites to be resolved based on their drift times. In the experiments described in this technology brief, nefazodone was incubated with rat hepatocytes and analysed using both DDA and IMS-enabled DDA. Co-eluting metabolites assigned as the glucuronides of dihydroxylated metabolites were characterised and the results processed, interrogated, and visualized using the UNIFI Scientific Information System.
Nefazodone (10 µM) was incubated with cryopreserved rat hepatocytes and relevant cofactors at 37 °C for 0, 15, 30, 60, 120, and 140 minutes. The incubations were terminated by addition of an equal volume of ice cold acetonitrile, centrifuged, and the supernatant submitted for analysis.
|
System: |
ACQUITY UPLC I-Class (FTN) |
||
|
Column: |
ACQUITY UPLC HSS T3, 1.8 μm, 2.1 x 50 mm |
||
|
Run time: |
4 minutes |
||
|
Vials: |
Waters Maximum Recovery |
||
|
Column temp.: |
45 °C |
||
|
Sample temp.: |
8 °C |
||
|
Injection volume: |
1 μL |
||
|
Flow rate: |
0.65 mL/min |
||
|
Mobile phase A: |
water + 0.1 % formic acid |
||
|
Mobile phase B: |
acetonitrile + 0.1 % formic acid |
|
Time |
%A |
%B |
Curve |
|---|---|---|---|
|
0 |
98 |
2 |
– |
|
2 |
0 |
100 |
6 |
|
3 |
0 |
100 |
6 |
|
3.1 |
98 |
2 |
6 |
|
4 |
98 |
2 |
6 |
|
MS system: |
Vion IMS QTof (operated under Driver 2.0) |
||
|
Ionisation mode: |
ESI+ |
||
|
Source temp.: |
120 °C |
||
|
Desolvation temp.: |
450 °C |
||
|
Desolvation gas: |
800 L/hr |
||
|
Capillary voltage: |
0.6 kV |
||
|
Cone voltage: |
40 V |
||
|
Reference mass: |
Leucine enkephalin [M+H]+ m/z 556.27658 |
|
Acquisition range: |
m/z 50–1200 |
||
|
Scan time: |
0.1 s |
|
Trigger MS/MS acquisition on: |
Include list (containing m/z of expected metabolites) Figure 1 |
|||
|
Acquire MS/MS when intensity exceeds: |
1000 detector counts |
|||
|
Maximum simultaneous MS/MS acquisitions: |
2 |
|||
|
Stop MS/MS: |
until time out (1 s) |
|||
|
Collision energy: |
Mass dependent ramp: Low 15-35 eV, High 20-50 eV. |
Pathway profiling with DDA/IMS-enabled DDA – UNIFI v1.8.2
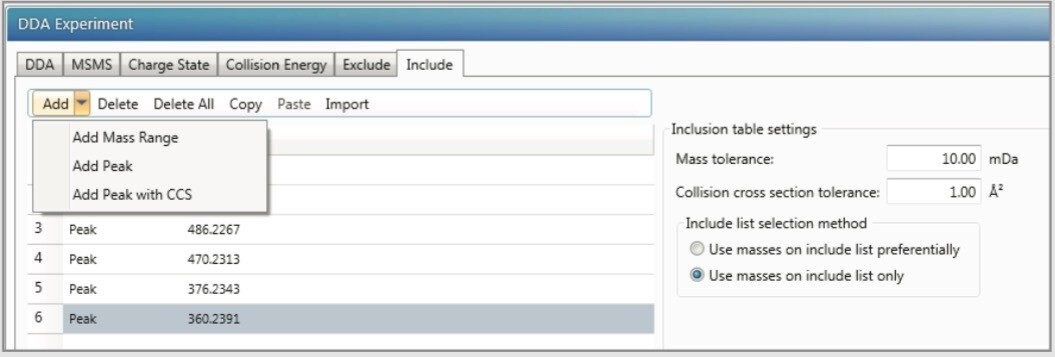
In this study, nefazodone and its metabolites were used as a model system to explore the advantages of IMS-enabled DDA using Vion IMS QTof Mass Spectrometer. Although data independent acquisition strategies such as MSE and HDMSE have been broadly adopted for discovery qualitative analysis, DDA remains an important tool in the MS toolbox, providing MS/MS spectra through quadrupole mass selection.1 IMS-enabled DDA affords greater selectivity than DDA since the former allows precursor selection using both m/z and drift time (Figure 2). This leads to more definitive MS/MS spectra.
The metabolic fate of nefazodone is well-characterized. Clayton et al., have shown that at least two glucuronides of dihydroxylated metabolites elute at approximately the same retention time, and have two distinct drift times.2 Using traditional MS-based DDA, it is not possible to resolve the spectra from these two metabolites since the MS/MS spectrum is a mixture of the product ions from a precursor ion derived from two different chemical entities. Figure 3 shows the precursor ion MS spectra and the product ion MS/MS spectrum of the glucuronide of dihydroxylated metabolites using DDA.

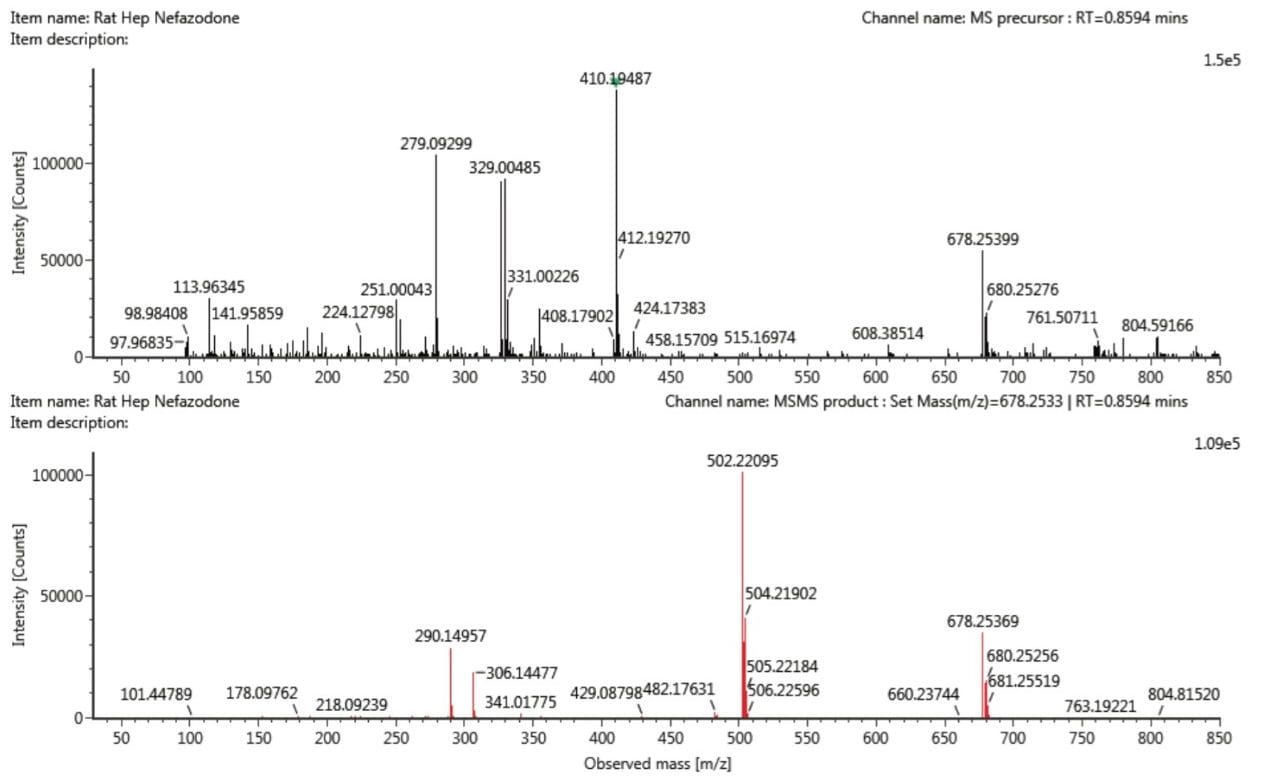
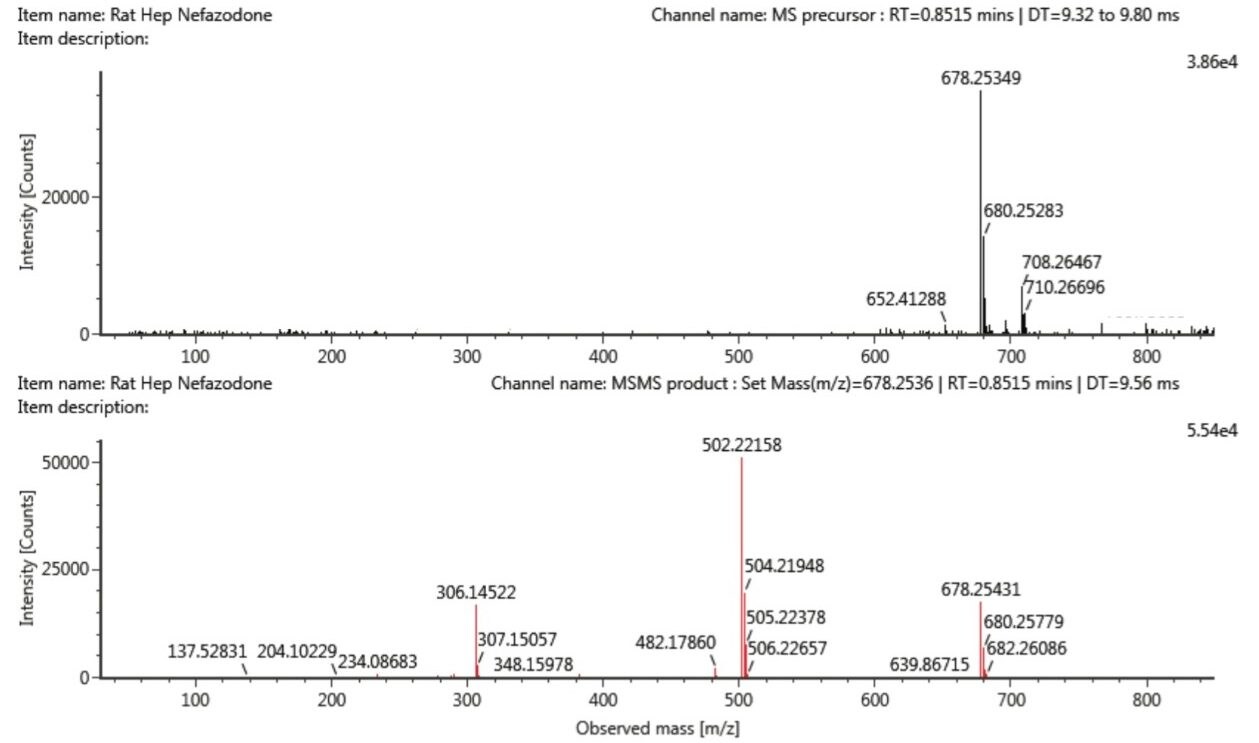
The same samples were analysed using IMS-enabled DDA, resulting in product ion MS/MS spectra for each of the co-eluting glucuronides of dihydroxylated metabolite, resolved on the basis of their two distinct drift times. The two spectra are shown in Figure 4 and highlight the differences in fragmentation between the two metabolites. The product ion assignments have been previously discussed (Clayton et al.) with the ion at m/z 678.25 attributed to the precursor ion. The product ion at m/z 502.22, a loss of 176 Da, is indicative of loss of the glucuronide moiety. Differences between the spectra were observed at lower mass, with fragment ions at m/z 306.14 in spectra for the metabolite with DT 9.56 msec and m/z 290.15 in the spectra for the metabolite with DT 10.13 msec, a difference of 16 Da, attributed to a hydroxyl group, thus aiding in product ion assignment. It was not possible to resolve these spectra from the two metabolites under traditional MS-based DDA (Figure 3).
Shown in Figures 5 and 6 are the precursor ion MS spectra and the product ion MS/MS spectra for the two metabolites. The precursor ion MS scan, acquired with IMS enabled, generates a much cleaner spectrum compared to traditional DDA (c.f. Figure 3). This highlights how using drift time for precursor selection can remove matrix interference in a complex sample, easing the burden of spectral interpretation.


Co-eluting metabolites attributed to glucuronides of dihydroxylated metabolites were successfully characterised using IMS-enabled DDA, generating two distinct precursor ion MS spectra and product ion MS/MS spectra for the drift time separated metabolites. The m/z and drift time filtered data provide cleaner, unambiguous spectra and increases confidence in structural assignment compared with simple m/z-selective DDA.
720005901, January 2017