
This application note explores the use of routine screening with UPLC and ion mobility to identify multiple protonation sites and different fragmentation patterns within the fluoroquinolone class of antibiotics.
Across many application areas, the applicability of ion mobility to small molecule analysis continues to increase, along with the understanding of how this technology can help address current analytical challenges. The reason, challenge, and methods of achieving successful fluoroquinolones analysis were briefly discussed in a previous application note, where we described the use of the ACQUITY UPLC I-Class System combined with ion mobility mass spectrometry to show how fluoroquinolone class of compounds can form protomers.1-5 Identification and characterization of the protomers of fluoroquinolones can now be routinely screened for using Waters UNIFI Scientific Information System. The software within UNIFI allows for the routine characterization of fragmentation pathways of the respective protomers to be visualized. In addition, it is possible to see the direct impact of the matrix upon protomer formation, and hence obtain a greater INSIGHT of the challenges of using MRM to perform residue analysis of fluoroquinolones.
Fluoroquinolones are a family of synthetic broad-spectrum antimicrobial agents that have been administered to livestock for different purposes, including the prevention and control of infections and for growth promotion. Due to concerns regarding the spread of resistant microorganisms in the human population, the U.S. Food and Drug Administration (U.S. FDA) introduced a ban on the use of enrofloxacin and ciprofloxacin in livestock production in September, 2005.7-9 The use of antibiotic growth promoting agents (AGPs) in animal husbandry has been forbidden in the European Union (EU) since 2006.10
This application note explores the use of routine screening with UPLC and ion mobility to identify multiple protonation sites and different fragmentation patterns within the fluoroquinolone class of antibiotics. It can be used as an important method development tool to support the unequivocal identification of fluoroquinolone antibiotics in crude tissue extracts. UPLC and ion mobility have been utilized to analyze crude extracts of porcine muscle tissue to determine the presence of antibiotic residues including the fluoroquinolone class.
The enhanced peak capacity provided by the combination of UPLC and ion mobility separation offers some unique advantages for profiling complex matrices. It uses a combination of high resolution mass spectrometry and high efficiency ion mobility-based measurements and separations. Ion mobility spectrometry (IMS) is a rapid, orthogonal, gas phase separation technique that allows another dimension of separation to be obtained within an LC timeframe. Compounds can be differentiated based on size, shape, and charge. In addition, both precursor ion and fragment ion information can be acquired in a single acquisition for all components.
A collision cross section (CCS) value is a robust and precise physicochemical property of an ion. CCS is an important distinguishing characteristic of an ion which is related to its chemical structure and three-dimensional conformation, where the shadow of a rotating three-dimensional ion, shown in Figure 1, represents the average collision cross section. Using CCS measurements can increase targeted screening specificity. CCS measurements generated have been entered into a scientific library within UNIFI. This allows the expected and determined CCS values to be utilized in order to screen and confirm fluoroquinolone protomer formation. Here we present CCS values (derived from ion mobility drift times) as a new identification parameter, which can distinguish protomers.
|
Analytes: |
Standards fluoroquinolones |
|
Extracts: |
Porcine tissue |
|
UPLC system: |
ACQUITY UPLC I-Class |
|
Column: |
ACQUITY UPLC BEH C18 1.7 μm, 100 mm x 2.1 mm |
|
Column temp.: |
40 °C |
|
Flow rate: |
0.6 mL/min |
|
Mobile phase A: |
Water (0.1% formic acid) |
|
Mobile phase B: |
Acetonitrile (0.1% formic acid) |
|
Injection volume: |
10 μL |
|
Time (min) |
%A |
%B |
|---|---|---|
|
Initial |
0.0 |
5.0 |
|
1.00 |
95.0 |
5.0 |
|
8.00 |
5.0 |
95.0 |
|
9.00 |
95.0 |
5.0 |
|
MS system: |
SYNAPT G2-S |
|
Ionization mode: |
ESI+ |
|
Capillary voltage: |
2.0 kV |
|
Cone voltage: |
25 V |
|
Desolvation temp.: |
550 °C |
|
Reference mass: |
Leucine enkephalin [M+H]+ = 556.2766 |
|
Acquisition range: |
50 to 1200 Da |
|
Acquisition rate: |
4 spectra/sec |
|
Collision energy: |
15 to 45 eV |
|
IMS T-Wave velocity: |
900 m/s |
|
IMS T-Wave pulse height: |
40 V |
|
IMS duty cycle: |
10.8 ms |
|
Drift gas: |
N2 |
Extracts of porcine muscle tissue were kindly provided by RnAssays BV for the purposes of this study. Briefly, known blank porcine muscle was fortified with 25 different antimicrobial compounds (from fluoroquinolone, tetracycline, and macrolide classes) at levels the relevant to the EU MRL concentrations prior to extraction. Tissue samples were mechanically homogenized in the presence of an aqueous/organic extraction solvent followed by a centrifugation step. An aliquot of the supernatant was removed and placed in an autosampler vial for subsequent LC-MS analysis.
For the assay performed, MassLynx MS data were acquired and processed with the UNIFI Scientific Information System, allowing ion mobility data to be processed in a conventional workflow for non-targeted accurate mass screening applications.
UPLC ion mobility MS has been explored as an important method development tool to support the unequivocal identification of fluoroquinolone antibiotics in crude tissue extracts. With UNIFI, it has been possible to routinely identify and characterize protomers of nine fluoroquinolones standards in a routine screening workflow.10 From the solvent standards analyzed, estimated CCS values of protomers formed for each fluoroquinolone were determined. The CCS values obtained have been incorporated into the UNIFI Scientific Library, which enabled the targeting of protomers.
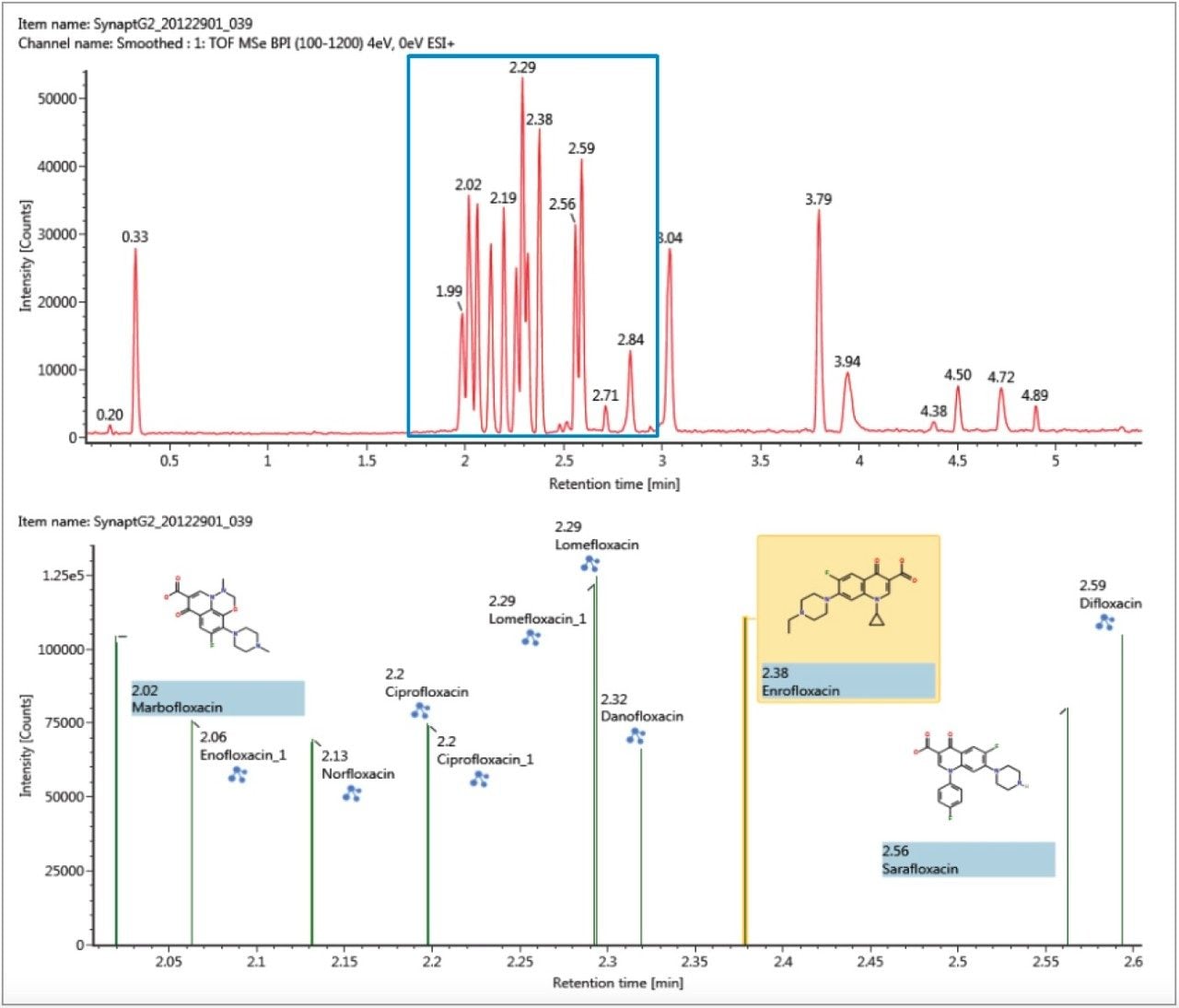
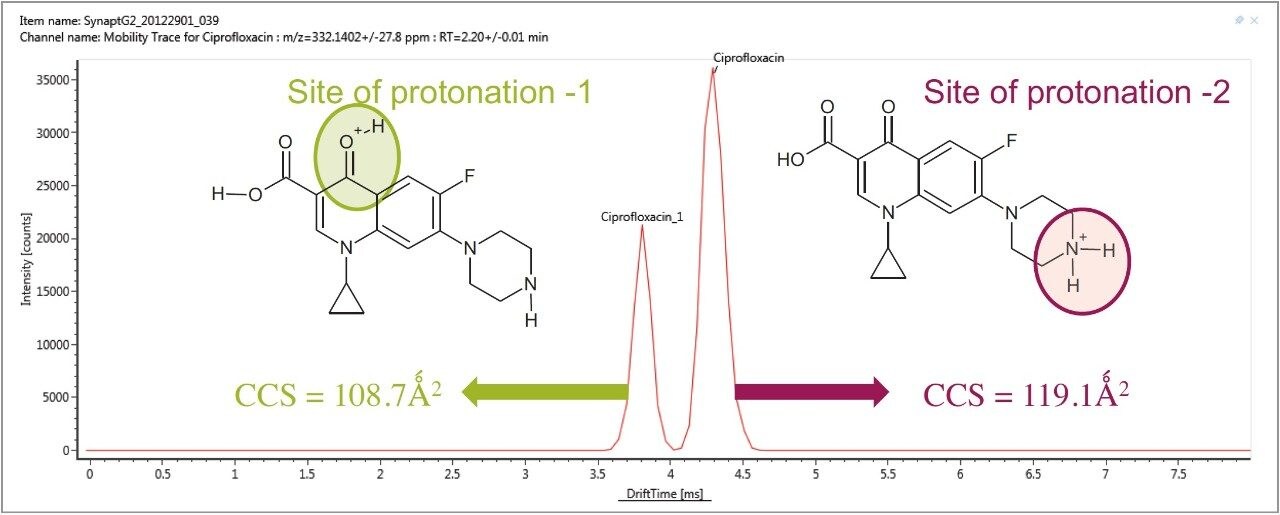
The antibiotic ciprofloxacin was determined to elute at retention time 2.19 min using the generic gradient conditions employed. Figure 2 shows the base peak ion chromatogram with the UNIFI Component Plot Summary for nine of the identified fluoroquinolone antibiotics, which eluted in the region highlighted in the base peak intensity chromatogram. From review of the data using the Component Drift Plot Summary, 18 fluoroquinolone species were identified. Each fluoroquinolone is comprised of two protomers, i.e. protonation at two different sites on the molecule that have been mobility separated, as shown in Figure 3. Each retention time shows two dots on the Component Summary Drift Plot, which indicate two forms of each fluoroquinolone. The two protomers of ciprofloxacin have been highlighted. The CCS values determined are presented in the Component Summary table of Figure 4. The functionality illustrated is unique to the UNIFI Scientific Information System.
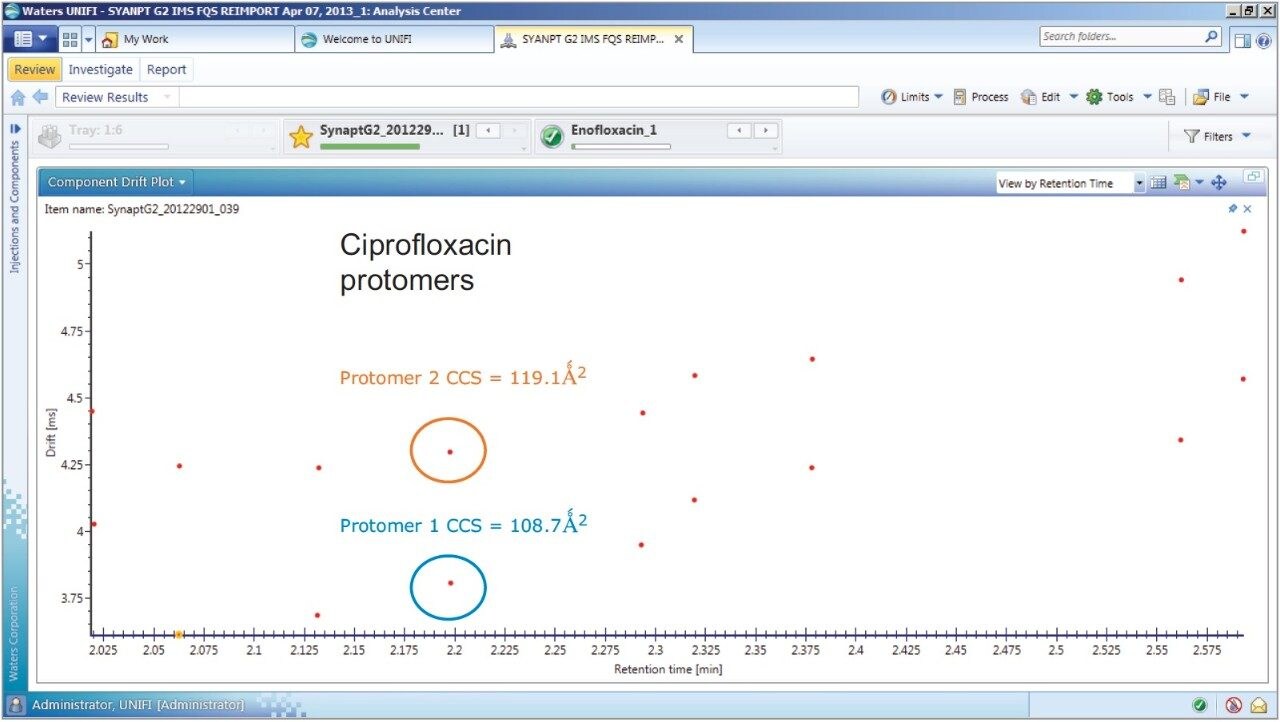
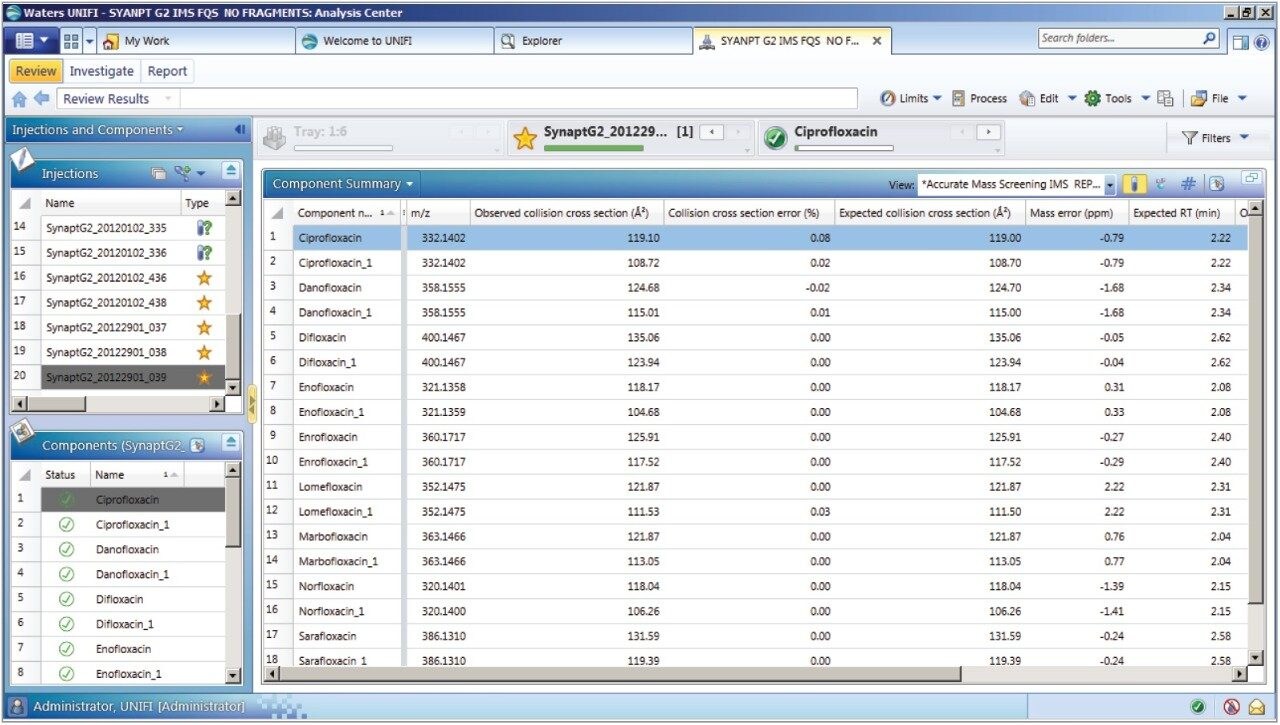
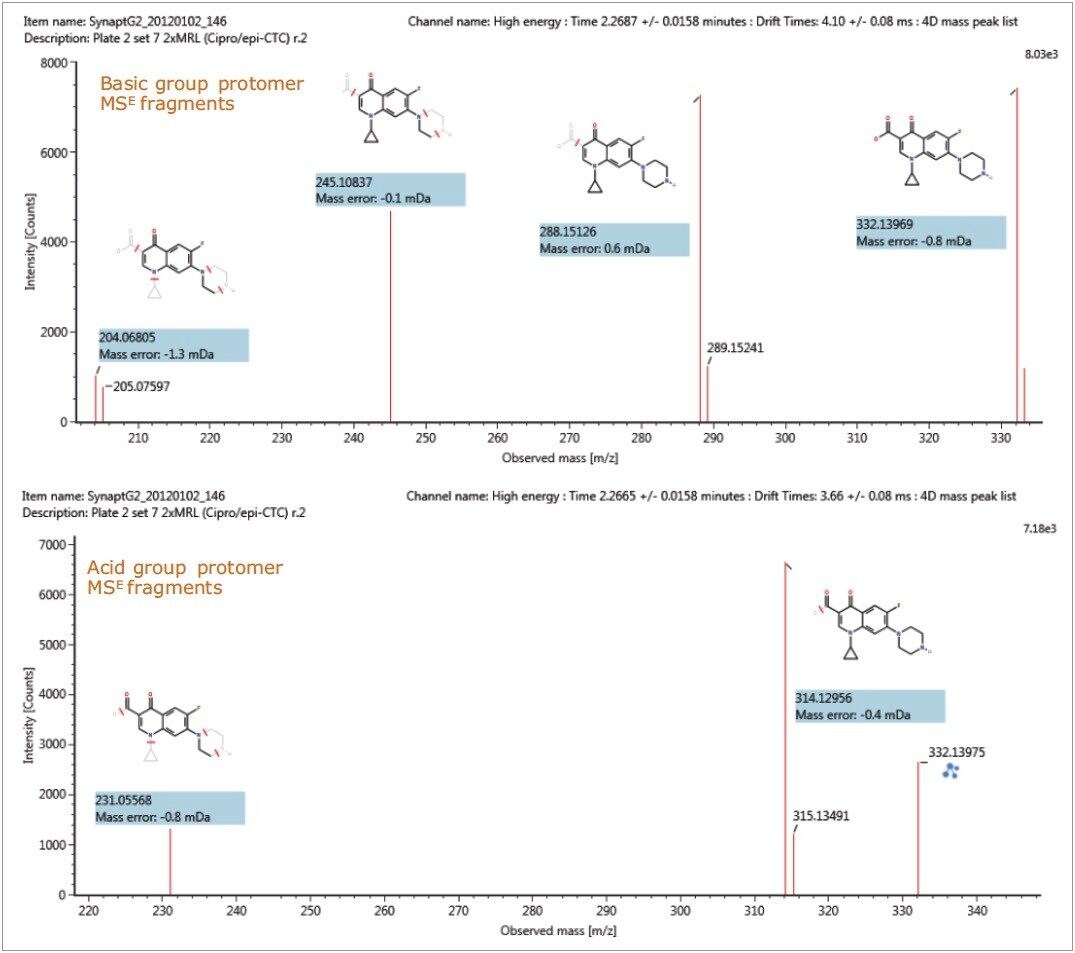
These protomer gas phase components, although they only differ with the site of protonation, have different collision cross sections. The example of ciprofloxacin is shown in Figure 5, and in this case a difference of >10 Å2 (angstrom is a unit of length equal to 10–10 m, one ten-billionth of a meter), was observed in this ion mobility study.11 For all fluoroquinolone protomers pairs observed, the respective difference between CCS pair values varied between 6 Å2 and 12 Å2 with respect to the protomer pairs. The mobility separation achieved enabled individual precursor ion and fragments of all nine fluoroquinolones to be obtained in one analysis. From a single component fragmentation spectra it was possible to determine that for ciprofloxacin, the two mobility separated species resulted from protonation taking place either on the acidic or the basic group. From method development with standards, specific CCS information was generated providing further specific information to be entered into the UNIFI scientific library. Using this information, veterinary drug residues can now be identified based on retention time, accurate mass, fragments, and CCS values. Ciprofloxacin’s estimated CCS values of 108.7 Å2 and 119.1 Å2 have been determined. Ciprofloxacin fragments at m/z 314 and m/z 231 are shown in Figure 6. These fragments are hypothesized to form from a species where ionization has taken place on the acidic group. Fragments observed at m/z 288 and m/z 245 resulted from protonation of the basic group.
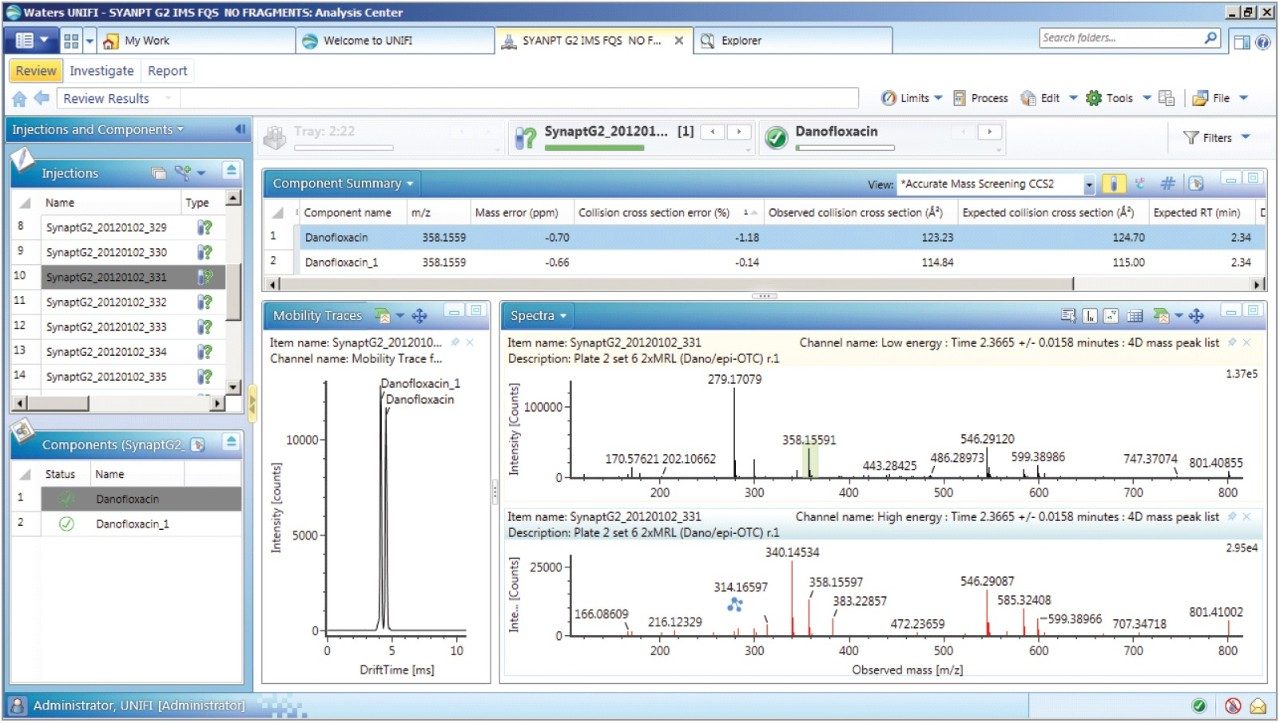
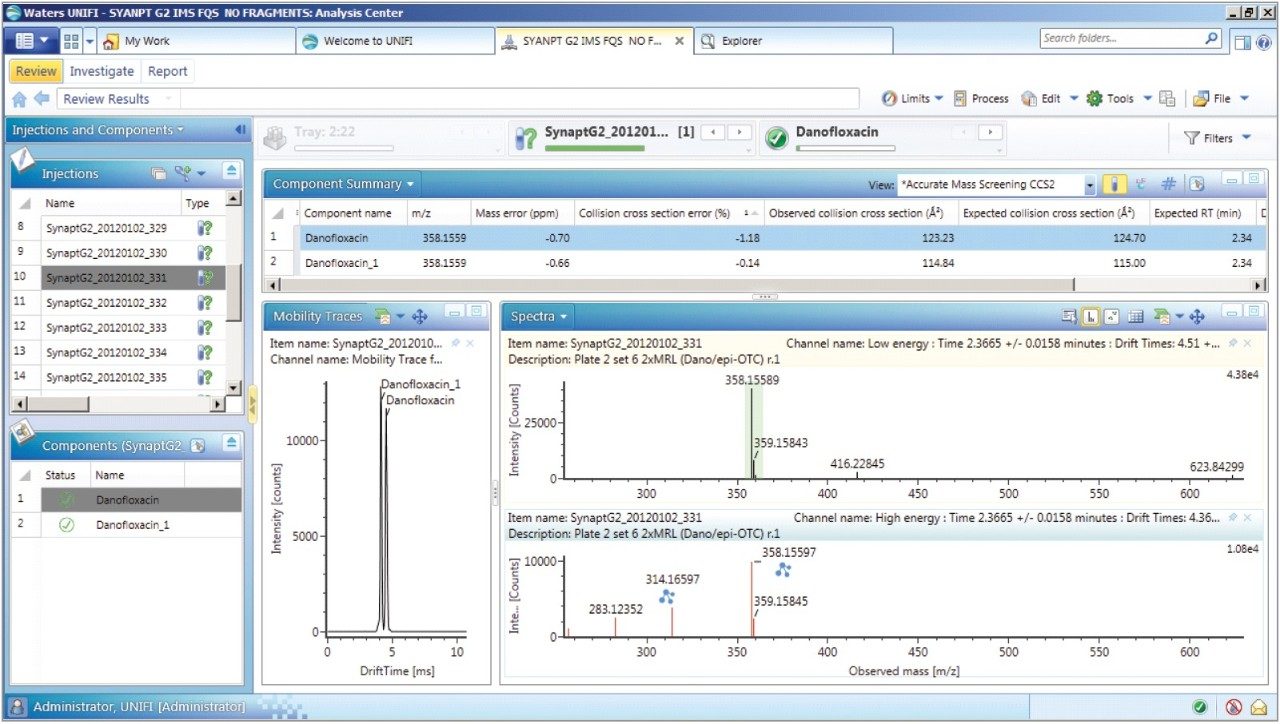
Once the CCS values and fragments of the individual fluoroquinolones were entered into the UNIFI scientific library, a series of spiked porcine extracts were screened to determine the presence of fluoroquinolones. Examples of the screening results are shown in Figures 7 and 8, where the identification of two protomers of danofloxacin in porcine extract are presented. Here, the benefits of ion mobility resolution and the functionality of UNIFI are demonstrated, showing the resolved protomers, as well as the removal of the matrix background from the identified component danofloxacin. It can also be seen that for both protomers, mass accuracy <1 ppm was obtained, and that the CCS error was within 2% of the expected CCS values (124.7 Å2 and 115.0 Å2). The observed retention time was 2.36 min and the individual protomer precursor ion/fragmentation spectra have been obtained. This data further illustrates how confidence in true identifications can be increased when using UPLC ion mobility in conjunction with the functionality available within UNIFI Software.
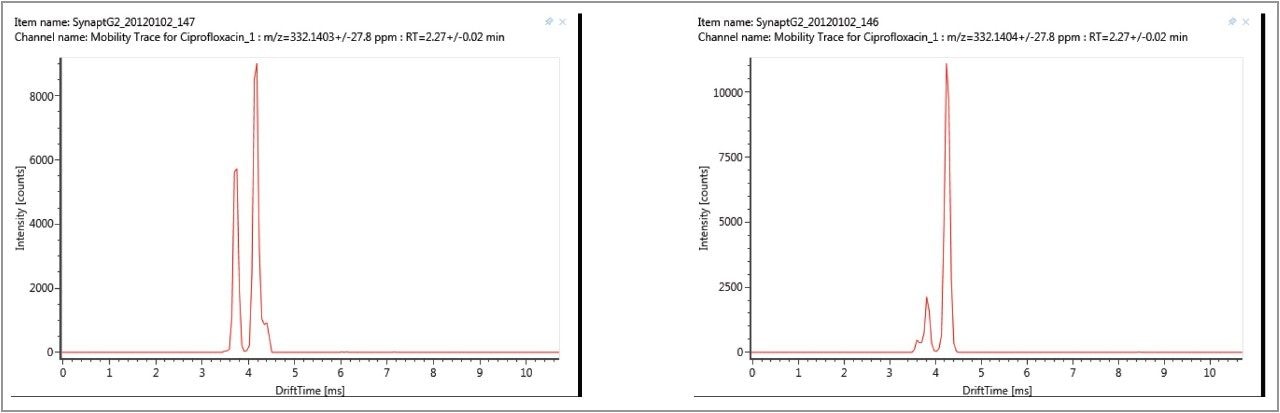
The impact of the matrix upon the gas phase intra-molecular protonation for ciprofloxacin is shown in Figure 9. The ability to routinely process ion mobility data within a workflow has made it possible to clearly observe fluctuations in the ratios of the fluoroquinolone protomers formed. As discussed, each protomer generates specific fragments, and from this data, an understanding can be obtained of why fluctuations in observed ion ratios can occur when monitoring MRM transitions.
Multiple protonated species have been observed for the fluoroquinolone antibiotics screened. The extent of the protonation multiplicity and its experimental variation is still being investigated. This data confirms that further consideration should be given to method development and the means of analysis chosen, since the ratio and formation of the protomers can vary with the eluent flow rate, capillary voltage, cone voltage, and matrix.
If MRM is the method of choice, consideration of the experimental conditions used and the specific transitions selected is imperative. The data presented illustrate that consistency in MRM transitions in inter/intra laboratory studies could easily be misinterpreted within and between different laboratories, and demonstrates the challenges of achieving reproducible results for these types of compounds. Ion mobility can provide a valuable tool for method development in order to ensure method robustness and consistency of results.
The benefits of UPLC ion mobility mass spectrometry can be demonstrated over traditional ‘shape selective’ ion mobility-based separation techniques, such as Field Asymmetric Waveform Ion Mobility Spectrometry (FAIMS). FAIMS is typically used to transmit only ions of a particular mobility, essentially acting as a filter. Using the ACQUITY UPLC I-Class System with HDMS, the fastest, ion mobility separation can be performed for all components regardless of the sample complexity, maximizing the duty cycle of the analysis and the amount of information obtained. Having a compatible duty cycle available also ensures spectral integrity is retained.
The benefits of time-of-flight mass spectrometry and historical data review (retrospective data analysis) is well known. Such historical data review is also required with ion mobility mass spectrometry. The discovery and presentation of multiple sites of protonation occurring during analysis for fluoroquinolone antibiotics can only be possible if the ion mobility data is acquired for all of the components in a sample. Continued development of the UNIFI platform’s functionality has enabled routine screening using ion mobility mass spectrometry, facilitating the opportunity to develop more reproducible, repeatable, and robust assays for a wide range application areas.
Waters kindly acknowledges Aldert Bergwerff and Wouter de Keizer (RnAssays) for the provision of the samples analyzed.
720005078, September 2017