
For research use only. Not for use in diagnostic procedures.
Consistent, reliable, and rapid identification and quantification of hundreds of lipid species can now be performed in a single run by the application of this workflow. This application note describes a tandem-quadrupole-based workflow that enables a greater understanding of the global lipidome and its association with diseases.
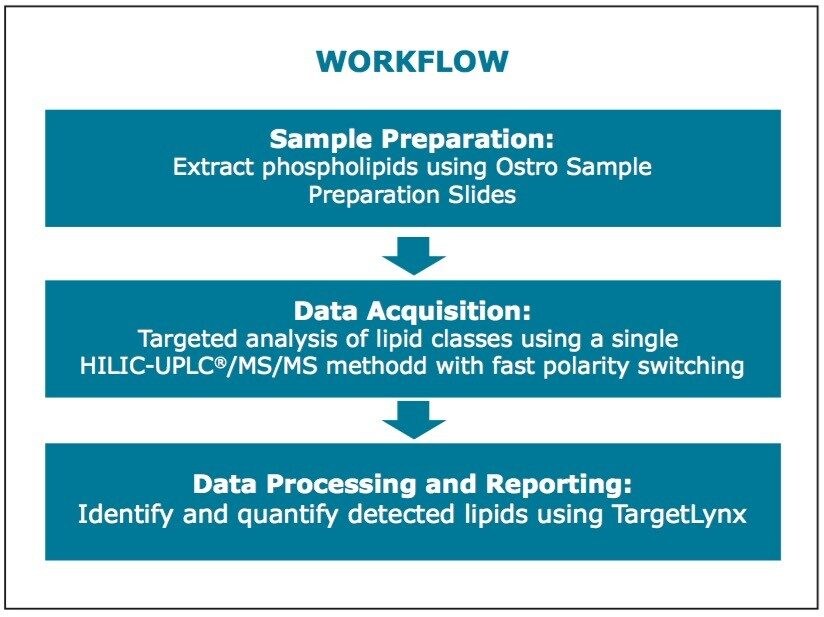
Large-scale quantitative and comparative lipidomic studies require the use of simple and high throughput workflows. To answer this need, a tandem quadrupole-based workflow has been developed. The benefits of this targeted lipidomics workflow include the following:
Lipids play many important roles in maintaining homeostasis of living organisms. Lipidomics analyses could further our understanding of mechanisms of disease, including the identification of biomarkers and potential drug targets.
Biofluids such as plasma are typically complex, with large lipid diversity across many orders of concentration. These, together with the chemical complexity of lipids, present demanding analytical challenges ranging from the sample preparation stage to the analytical techniques used to identify and quantify key lipids. Today, many variations of the Bligh and Dyer method are used for total lipid extraction and purification, with equal amounts used for mass spectrometric analysis.
Recent advances in lipidomics have made use of developments in chemistries and instrumentation, most notably the use of off-line enrichment or solid-phase sample preparation products1,2 and the coupling of UltraPerformance LC with mass spectrometry. However, there is little standardization across platforms and workflows for a complete analysis.
Presented here is a tandem quadrupole-based phospholipid analysis workflow from extraction to separation, identification and quantification of the phospholipids from a single vendor. These commercially available products packaged as a complete solution are provided to ease the strains and increase the productivity of laboratories undertaking longitudinal studies spanning hundreds of lipids over thousands of samples.
|
System: |
ACQUITY UPLC |
|---|---|
|
Column: |
ACQUITY UPLC BEH HILIC 2.1 x 100 mm, 1.7 μm |
|
Column temp.: |
30 °C |
|
Mobile phase A: |
95:5 acetonitrile/water with 10 mM ammonium acetate, pH 8.0 |
|
Mobile phase B: |
50:50 acetonitrile/water with 10 mM ammonium acetate, pH 8.0 |
|
Gradient: |
0% to 20% B for 10 min |
|
Flow rate: |
500 μL/min |
|
Injection volume: |
3.0 μL, partial loop |
|
Mass spectrometer: |
Xevo TQ-S |
|
Ionization mode: |
ESI, +/- switching |
|
Capillary voltage: |
3.8 kV (+) / 1.9 kV (-) |
|
Desolvation temp.: |
450 °C |
|
Desolvation gas: |
1000 L/h |
|
Source temp.: |
150 °C |
|
Collision cell pressure: |
3.6 x 10-3 mBar |
Human plasma samples were obtained from the Centre for Life Sciences, National University of Singapore. The protocol described here follows a recently published application note.3
100 μL of human plasma was loaded into each well of a Waters Ostro Sample Preparation Plate fitted onto a vacuum manifold. 800 μL of ethanol was added to each well and mixed thoroughly by aspirating the mixture 10x using a micropipette. A vacuum of approximately 15” Hg was applied to the plate until the solvent was completely drained. This step was repeated with another 800 μL of ethanol with the total fraction collected labelled as the “flow through.”
800 μL of elution solvent (4.5:4.5:1.0 chloroform/methanol/triethylamine) was added to each well, and the fraction was collected under 15” Hg vacuum as the “eluate.” This step was repeated, bringing the total fraction volume to approximately 1600 μL.
Both the eluate and flow through fractions were dried down under nitrogen, and reconstituted with 200 μL 1:1 (v/v) chloroform/methanol. 1 μL of the eluate fraction was combined with 99 μL of the flow through fraction to give a 1:100 dilution. This combined sample was then injected into the UPLC/MS system.

While advancements in instrument technologies such as UPLC have enabled faster analysis of large numbers of samples, sample preparation time has been commonly viewed as the bottleneck.
When performed manually, lipid extraction using the Bligh and Dyer method takes approximately one hour and the Ostro plate method 1.5 hours. As the Ostro plate is available in a standard 96-well plate design, it can be easily adapted to most automated liquid handlers making the process time for one sample or 96 samples approximately the same, for example, 1.5 hours. Whereas, it took almost two days to manually process 96 samples using the Bligh and Dyer method.
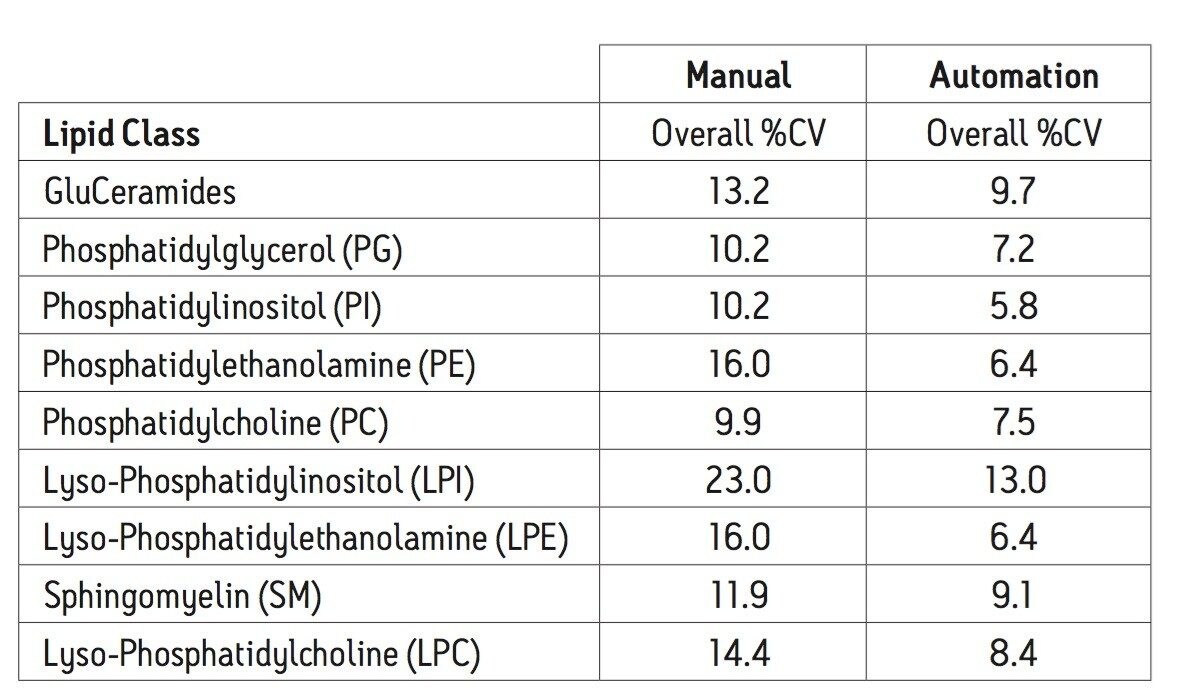
Using an automated liquid handler to process the plasma samples, as shown in Figure 2, the well-to-well reproducibility (%CV) improved compared to manually performing the extraction. This was true for all classes of lipids analyzed, with improvements ranging from 25% (SM) to 60% (PE).
In reversed-phase chromatography of lipids, separation is governed by lipophilicity, alkyl chain length, and degree of saturation for each individual lipid. These result in broad MRM acquisition time windows4 that, in turn, negatively affect the instrument’s duty cycle, thus hindering accurate quantification.
In the HILIC-UPLC/MS method used in this application note, there was a clear and reproducible separation of the various classes of lipids. This was observed very clearly by the difference in retention times of PCs and SMs, which are normally difficult to identify and quantify using reversed-phase methods.5, 6
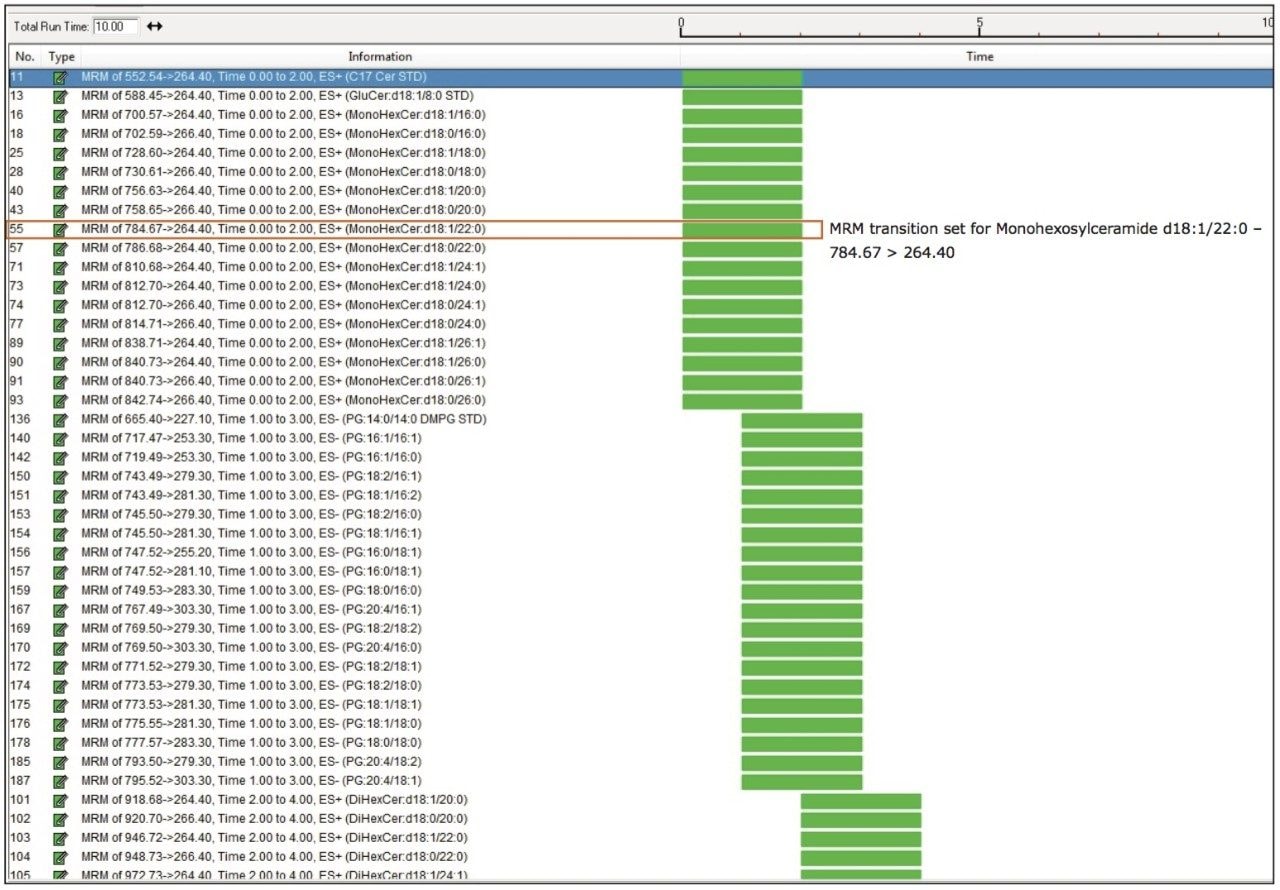
By leveraging the reproducible retention times of the lipid classes, MRM acquisition time windows were reduced to two minutes per class, as shown in Table 1. This allowed for the creation of a single MS method to analyze the combined “flow through” and “eluate” fractions in a single run. A total of 215 MRM transitions (+/- polarity) including internal standards were created in this method. Figure 3 shows each lipid transition set up as a single function, for example, monohexosylceramide d18:1/22:0, which will limit the addition or subtraction of lipids to only those of interest to the operator.

A complementary data processing method was created using the TargetLynx Application Manager, as shown in Figure 4. The insert shows how easily peak information can be “dragged and dropped” into the data processing method.
Once the processing method had been set with the appropriate transitions and retention times for each lipid, batch processing for any number of samples run under the same conditions described above can be performed. Figure 4 shows a typical TargetLynx results view. Using monohexosylceramide d18:1/22:0 as an example, the application manager automatically integrates both the sample peak and the pre-defined internal standard (IS) peak and calculates the concentration of the lipid in the sample against the known spiked concentration of the IS. A user-defined report can then be printed, or these results can be exported into a number of popular generic formats for further statistical analysis.

Consistent, reliable, and rapid identification and quantification of hundreds of lipid species can now be performed in a single run by the application of this workflow. The high throughput nature of the workflow utilizing automation technologies and automated data processing and reporting means that large-scale comparative lipidomic studies can be routinely used by laboratories around the world. In addition, the consistency obtained from this standardized platform means that data can be shared and compared across various sites, thereby enabling a greater understanding of the global lipidome and its associations with diseases.
720004521, December 2012