
For research use only. Not for use in diagnostic procedures.
This application note illustrates the value of UPLC-MS when applied to a blind study of urine samples from 25 patients with five unknown inborn errors of metabolism (IEM).
UPLC-MSE was used to profile diagnostic metabolites in a single analytical run. This approach enables the simultaneous unveiling of more subtle changes in the urinary metabolite profiles of inborn errors of metabolism (IEM) patients, which may contain information needed to obtain biologically relevant conclusions. Use of this metabolic profiling approach allows for the detection of previously unobserved markers, providing wider metabolic insight into disease processes.
LC-ESI-MS is currently the most prevalent separation technique employed in neonatal screening, used to diagnose over 30 inborn errors of amino acid, organic acid, and fatty acid metabolism. Recent analytical improvements in this approach have reduced the false-positive result rate, as compared to other conventional techniques.1,2 However, UPLC-MS has not yet been used for analyzing inborn errors of metabolism. UPLC uses columns packed with sub-2 μm particles, resulting in better chromatographic resolution, excellent separation, and increased sensitivity than conventional high performance liquid chromatography (HPLC).3,4 The combination of quadrupole time-of-flight (Q-Tof) mass spectrometers with UPLC systems provides the power of accurate mass measurements with fragmentation information, and has proved successful for the identification and structural elucidation of metabolites in complex biofluids.5 This application note illustrates the value of UPLC-MS when applied to a blind study of urine samples from 25 patients with five unknown inborn errors of metabolism (IEM). A 12 min UPLC-QTof/MS assay was developed for the targeted profiling of organic acids using an ACQUITY UPLC C18 BEH, 2.1 x 100 mm, 1.7 μm Column, but was designed to also provide information on other classes of urinary metabolites. All five IEMs were identified correctly, and could be differentiated through principal component analysis (PCA) from data generated in MarkerLynx XS Application Manager. All organic acids were identified through a combination of MSE and MS/MS and compared with authentic standards. This work illustrates the capability of UPLC-MS for the rapid and accurate diagnosis of inborn errors of metabolism.
Data were acquired using UPLC-MSE, a comprehensive data collection technique that enables collection of precursor and product ion information for virtually every component of a mixture. Exact mass measurements were acquired using leucine enkephalin (MW= 555.62) as a single point lockmass to ensure accuracy and reproducibility; a 200 pg/μL leucine-enkephalin in H2O:ACN (50:50) solution was used at a flow rate of 30 μL/min. Data were collected in centroid mode, the lockspray frequency was set at 5 seconds, and data were averaged over 10 scans. The mass spectrometer was calibrated across the mass range 50 to 1000 Da using sodium formate.
25 urine samples were collected from five groups of patients with unknown IEM. A 150 µL aliquot of each urine sample was added to 150 µL of water. The samples were centrifuged at 16,089 g for 10 minutes to remove particulates, and 250 µL of diluted urine was transferred into 96-well plates.
|
System: |
ACQUITY UPLC |
|
Column: |
ACQUITY UPLC C18 BEH, 2.1 x 100 mm, 1.7 μm |
|
Column temp.: |
40 °C |
|
Sample temp.: |
4 °C |
|
Mobile phase A: |
Water, 0.1% formic acid |
|
Mobile phase B: |
Acetonitrile, 0.1% formic acid |
|
Flow rate: |
0.5 mL/minute |
|
Injection vol.: |
5 μL |
|
Time (min) |
% A |
Curve |
|---|---|---|
|
0.0 |
100 |
— |
|
1.0 |
85 |
6 |
|
3.0 |
50 |
6 |
|
6.0 |
5 |
6 |
|
9.9 |
5 |
6 |
|
10.1 |
100 |
6 |
|
12.0 |
100 |
6 |
|
MS system: |
Q-Tof Premier |
|
ESI: |
Positive and Negative |
|
Scan range: |
50 to 1000 Da |
|
Capillary voltage: |
3.2 Kv (pos mode), 2.4Kv (neg mode) |
|
Source temp.: |
120 °C |
|
Desolvation temp.: |
350 °C |
|
Cone voltage: |
35 V |
|
Desolvation gas flow: |
900L/hr |
|
Collision energy (CE): |
Low CE: 5 eV |
|
High CE: |
Ramp of 10 to 30 eV |
|
MS/MS experiments: |
CE 15 to 30 eV |
The data were acquired using Waters MassLynx Software, v.4.1 and were processed using MarkerLynx XS Application Manager. The software is designed to extract all features from the dataset and consequently enables metabolite detection, multivariate statistical analysis, data visualization, and reporting. Data were also exported to SIMCA-P from Umetrics for further analysis.
Using the developed UPLC-MSE method, 21 specific urinary metabolites were separated and detected over 12 minutes, as detailed in Table 1. Metabolite identifications were confirmed through the acquisition of UPLC-MSE data from authentic standards, followed by retention time and exact mass precursor, as well as product ion correlation. Of the 21 standards, 12 metabolites were detected in both ionization modes, eight in negative mode only, and one in positive mode only. This demonstrates the need to use both modes of ionization. The total analysis time of 12 minutes not only provided a rapid method for the targeted analysis of urinary organic acids, but also enabled the separation and detection of other urinary metabolites. This method also facilitated the elucidation of additional markers for these inborn errors of metabolism.
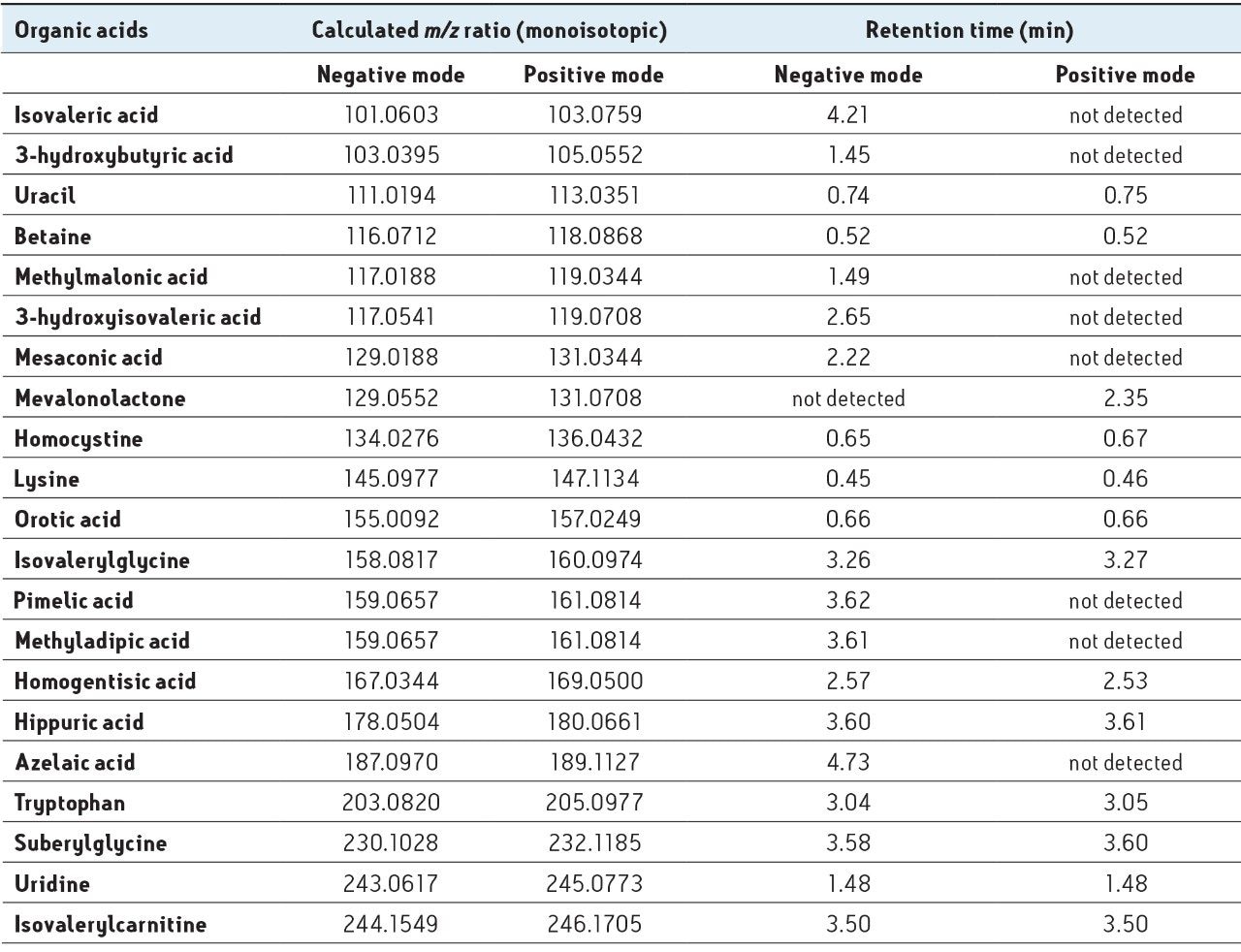
Negative and positive ionization mode base peak intensity (BPI) UPLC-MS chromatograms illustrate the metabolite-rich region between 0.4 to 6.0 min for each IEM, and the corresponding control urine sample, are shown in Figure 1. Although visual inspection yielded some obvious differences between the chromatograms, these represented a small proportion of total metabolic response, and the subtle differences contained within the chromatograms, were difficult to discern by eye. To conduct a manual comparison of the chromatographic response of each metabolite would be very labor-intensive and ultimately fruitless, as advanced normalization techniques are typically required to account for variations in sample concentration. In order to address this issue, data processing was performed with MarkerLynx XS Software, which offers a rapid and reliable method for analysis of complex metabolite data. Using MarkerLynx XS, data was extracted and collated to form a marker table. This table contains all the relevant information to perform statistical analysis and the data can be easily exported into SIMCA-P in order to perform additional multivariate analysis. For this study, PCA was used to analyze the data set.
Analysis of the urine, using data collected and analyzed in this manner showed elevated levels of specific metabolites related to five types of IEM. These IEM conditions were subsequently confirmed as:
A) methylmalonic aciduria (MMA)
B) mevalonic aciduria (MEV)
C) isovaleric aciduria (IVG)
D) alkaptonuria (ALK)
E) ornithine transcarbamylase deficiency (OTC)
The benefit of collecting data in both positive and negative ion modes is illustrated for isovaleric aciduria. In this case, two metabolites – isovalerylglycine [M+H]+ and isovalerylcarnitine [M-H]-, were detected and identified as potentially important markers. Due to the rapid analysis times of UPLC-MS, this type of complementary information can be obtained to provide additional disease confirmation in less time than it would take for a single HPLC run.
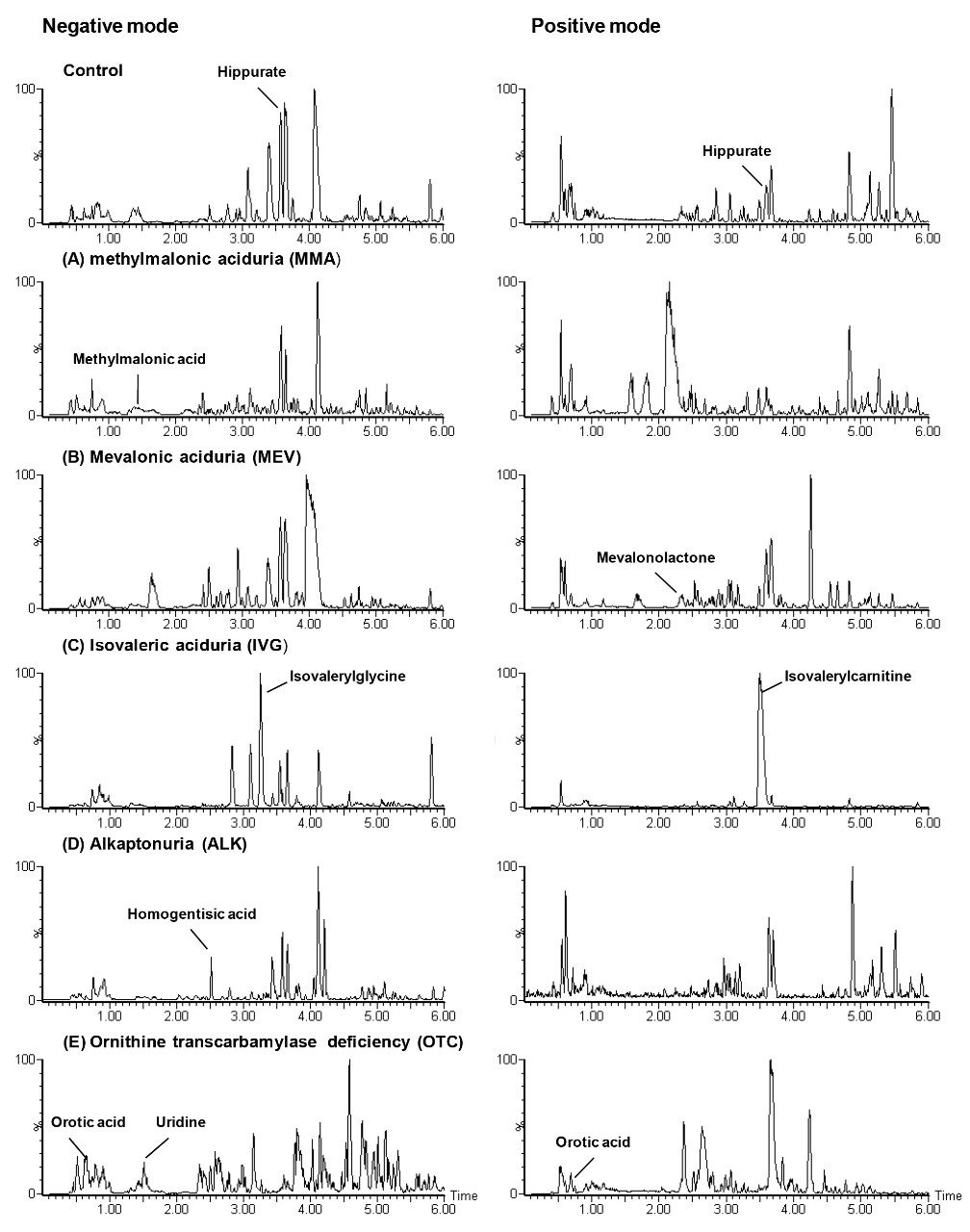
Figure 2 shows the PCA scores plot produced in SIMCA-P using data acquired in negative ion and processed through MarkerLynx XS. It can be seen that there is some differentiation between the different diseases, based upon the information submitted to the statistical analysis. This differentiation was particularly evident for MMA and MEV. From the scores plot, two samples show obvious outliers from the groups to which they belong. Further investigation of these samples concluded that they were obtained from patients who were undergoing treatment for an IEM disease. The corresponding loadings plot (not shown) provided confirmation of the discriminating markers as well as information on more subtle changes in the urinary metabolite profiles due to the specific diseases.
This application note demonstrates the applicability of a UPLC-MSE approach for the rapid and reliable collection of data for metabolomics studies and the utility of this approach to identify compounds that are descriptive of inborn errors of metabolism. Employing MarkerLynx XS for data processing and analysis provides a robust method for data extraction from complex samples, and allows the individual diseases to be distinguished readily. Using this UPLC-MSE approach, panels of diagnostic metabolites can be profiled in a single analytical run, without the need for disease-specific protocols, potentially increasing sample throughput. While known metabolite markers for each disease could be easily detected and identified, this approach enables the simultaneous unveiling of more subtle changes in the urinary metabolite profiles of these patients, which may contain much of the information needed to draw biologically relevant conclusions on IEM diseases. Importantly, despite some inborn errors of metabolism being well-characterized, there is still scope for new markers to be discovered, and due to its untargeted nature, the application of this metabolic profiling approach allows for the detection of previously unobserved markers, providing wider metabolic insight into disease processes. This robust technology allows for further characterization of IEMs in terms of urine and plasma metabolites, and could aid in the metabolic understanding of novel IEMs and related diseases. This in turn may facilitate more accurate or earlier disease diagnoses, as well as improvements in the treatment of these diseases through more timely therapeutic intervention.
720003942, April 2011