In this application note, a generic UPLC-MS assay that offers comprehensive HCP identification and quantification for biotherapeutic protein samples is described.
***nanoACQUITY UPLC applications readily transfer to the ACQUITY UPLC M-Class System***
Most biotherapeutics today are produced by recombinant DNA technology using a well-selected host cell system. Host cells express a large number of their own proteins that can easily contaminate the recombinant protein drug. Even after sophisticated purifications steps, low levels (1 to 100 ppm) of host cell proteins (HCPs) may still remain in the final purified biopharmaceutical product. Because HCPs can sometimes trigger an unpredictable immunogenic response, regulatory guidelines stipulate that they need to be identified and quantified to protect patient safety.
The presence or absence of HCPs in protein drugs can determine whether or not a biopharmaceutical is accepted by the regulatory agencies. For example, in 2008, the European Medicines Agency (EMA) approved a recombinant form of human somatropin only after the manufacturer added additional purification steps to remove the HCPs responsible for immunogenic response in patients.1 The same agency rejected an interferon biosimilar in 2006 because of insufficient validation for immunogenicity testing.
All analytical methods employed for measuring HCPs face significant challenges due to the wide dynamic range of the protein concentration (four to five orders of magnitude). Some widely used analytical methods, such as process-specific ELISAs and western blots,2 require prior knowledge regarding the nature of HCP contaminants. In addition, process specific immunoassays are both time consuming (e.g., six months), and expensive to develop (more than $100K), and are not readily adapted to fully evaluate biopharmaceutical products from different cell types and purification schemes.
Two-dimensional gel electrophoresis coupled to fluorescent staining,3,4 another popular method for HCP analysis, is only semi-quantitative, has limited dynamic range (two to three orders of magnitude) and requires additional, confirmatory techniques (e.g., mass spectrometry) for HCP identification. Although commercial ELISA kits are developed for generic application to the monitoring of HCPs, they are less specific than the process-specific immunoassays, and cannot offer a complete coverage for all the existing HCPs in the samples.5,6
An organization that can demonstrate that it is capable of accurately identifying and monitoring the HCPs in its biotherapeutics is more likely to overcome regulatory hurdles in the acceptance of its products.
In this application note, a generic UPLC-MS assay that offers comprehensive HCP identification and quantification for biotherapeutic protein samples is described. The assay applies an on-line two-dimensional LC approach for peptide separations and a high-resolution and high-mass-accuracy mass spectrometer for protein identification and quantification.
In contrast to the traditional 2D-chromatography setup schemes that are based on strong cation exchange (SCX) and low pH reversed-phase7 separation, the 2D method employed here couples a high pH reversed-phase (RP) separation to a low pH RP separation to achieve maximum chromatographic separation and to cope with the complexity and the wide dynamic range that are encountered in the HCP samples.
In addition, a multiplexed data acquisition method (MSE) is employed in the mass spectrometric analysis so low-abundance HCP peptides can be reproducibly sampled and identified without bias.
Furthermore, a fast quantitative assay has been developed based on multiple reaction monitoring (MRM) principles to provide a high-throughput method for monitoring HCP variation in samples from a variety of manufacturing/purification conditions.
In this application note, we evaluate the performance of the assay using monoclonal antibody samples derived from different purification methods.
|
2D-UPLC system: |
nanoACQUITY UPLC System with 2D Technology and on-line dilution |
|
Column: |
XBridge BEH300 C18, 5 μm, 1 x 50 mm, (p/n 186003615) |
|
Flow rate: |
10 μL/min |
|
Mobile phase A: |
20 mM ammonium formate in water (pH 10) |
|
Mobile phase B: |
Acetonitrile |
|
Step-elution gradient: |
A 10-step elution gradient to fractionate the peptides in the first dimension at pH 10 is undertaken. The percentage of mobile phase B in each step is: 10.8, 12.4, 14.0, 15.0, 16.7, 18.6, 20.4, 25.0, 30.0, and 50.0%, respectively. |
|
Diluting solution: |
0.1% TFA in Milli-Q water, 100 μL/min flow rate |
|
Trap column: |
Symmetry C18, 0.5 x 20 mm, 5 μm (2.7 μL internal volume) |
|
Column: |
ACQUITY UPLC BEH C18, 0.3 x 150 mm, 1.7 μm (p/n 186002605) |
|
Column temp.: |
65 °C |
|
Flow rate: |
12 μL/min |
|
Mobile phase A: |
0.1% FA in Milli-Q water (pH 2.4) |
|
Mobile phase B: |
0.1% FA in ACN |
|
Gradient elution: |
7 to 35% of mobile phase B in 30 min |
|
MS system: |
SYNAPT HDMS |
|
Acquisition time: |
0.5 s |
|
m/z range: |
50 to 1990 |
|
ESI spray voltage: |
2.6 kV |
|
Cone voltage: |
37 V |
|
Source temp.: |
120 °C |
|
Low-energy fragmentation: |
5eV (fixed) |
|
High-energy fragmentation: |
Collison Energy ramp between 15 and 35 eV |
|
LC system: |
ACQUITY UPLC |
|
Column: |
ACQUITY UPLC BEH300 C18, 2.1 x 150 mm 1.7 μm packed (p/n 186003687) |
|
Column temp.: |
35 °C |
|
MS system: |
Xevo TQ MS |
|
ESI spray voltage: |
3.5 kV |
|
Cone voltage: |
37 V |
|
Source temp.: |
90 °C |
|
MS1/MS2 isolation window: |
0.75 Da (FWHM) |
|
Dwell time: |
20 to 30 ms |
ProteinLynx Global SERVER (PLGS) 2.4
The MIX-4 protein digest standard was prepared by diluting stock solutions of the individual MassPREP protein digests of ADH (p/n 186002328), PHO (p/n 186002326), BSA (p/n 186002329), and ENL (p/n 186002325) in 20 mM ammonium formate, pH 10, to achieve a final concentration of 20 nM ADH, 4 nM PHO, 1 nM BSA, and 0.2 nM ENL.
A chimeric anti-phosphotyrosine IgG1 monoclonal antibody (PTG1 mAb) was expressed in two different Chinese hamster ovary (CHO) cell lines and purified by Protein A chromatography using two different protocols following manufacturer recommendations.
Among the six samples analyzed, four samples labeled as A1, B1, A2, and B2 were expressed in DG-44 CHO cells, while two samples labeled C and D were expressed in CHO-S cells. Two different purification protocols were followed. Samples A1/A2, and B1/B2 were biological replicates, grown under identical conditions.
Five protein standards (LA, PHO, ADH, BSA, and ENL) were spiked in 250 µL of PTG1 (5 to 10 mg/mL) and the resulting protein mixture was denatured with 0.1% RapiGest for 15 min at 60 °C, reduced with 10 mM DTT for 30 min at 60 °C, alkylated with 20 mM IAM for 30 min (at RT) and digested overnight (37 °C) with porcine trypsin (Promega) using a 1:20 (w/w, enzyme:protein) ratio. After digestion, the RapiGest surfactant was decomposed by adding 5 µL of pure TFA and the samples were incubated for 30 min at 37 °C and centrifuged (10 min at 10,000 rpm) to separate the insoluble component of RapiGest by precipitation. After adjusting the pH of the supernatant solution to pH 10 using a solution of 2 M ammonium formate (pH 11), the digest volume was brought to 1 mL using 20 mM ammonium formate (pH 10). The amounts of spiked protein digests loaded on-column using a 100 µL sample loop were: 4000 fmoles of LA (bovine alpha-lactoglobulin), 800 fmoles of PHO, 320 fmoles of ADH, 80 fmoles of BSA, and 16 fmoles of ENL.
For MRM quantification, three 13C15N-isotopically labeled peptides (Sigma Aldrich) were spiked at a concentration of 20 nM into 250 µL of PTG1 digest that was prepared following the digestion protocol described above.
A schematic diagram illustrating the operation of the 2D-UPLC system during sample loading, sample elution from the first dimension, and sample separation from the second dimension is shown in Figure 1A–C. Peptide samples are loaded under basic conditions (pH 10) on the first-dimension column (Figure 1A) and fractionated by RP chromatography using the 10-step elution with increasing acetonitrile concentrations (Figure 1B). Each peptide fraction is diluted (on-line) with a solution containing 0.1% TFA (pH 2.1) at a 1:10 ratio so the peptides eluted from the first dimension can be temporarily retained on a trapping column. The peptides are then separated on the second-dimension analytical column using a 30-min gradient under acidic conditions (Figure 1C).
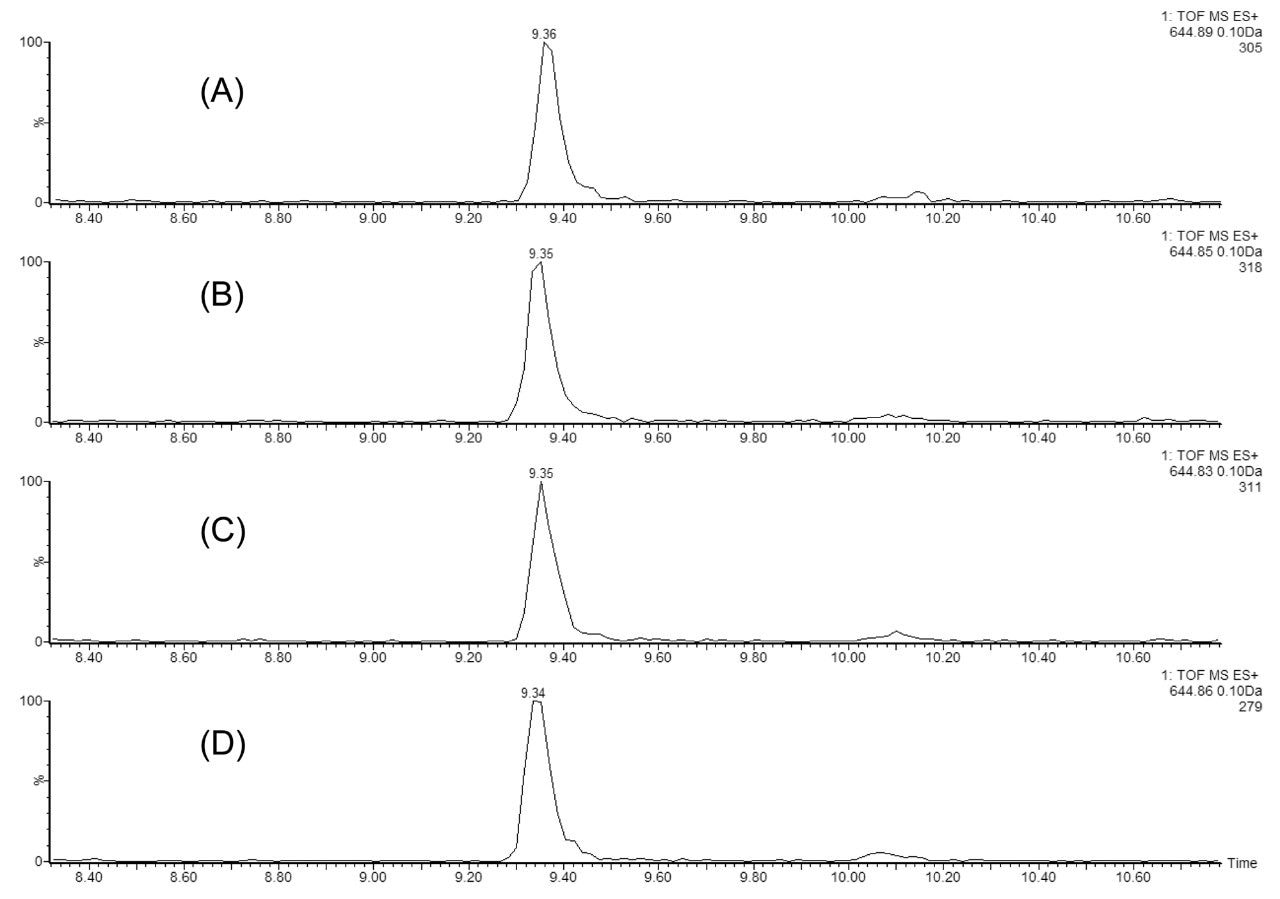
A critical chromatographic parameter in multidimensional chromatographic separations is the reproducibility of peptide fractionation during an extended period of operation. This is illustrated in Figure 2 using a MIX-4 protein digest, which contains 20 nM of ADH, 4 nM of PHO, 1 nM of BSA, and 0.2 nM of ENL in 20 mM ammonium formate (pH 10). Figure 2 shows the extracted mass chromatograms of the T43 peptide from ENL protein (VNQIGTLSESEIK, monoisotopic peak [M+2H]2+ of 644.86), from the second dimension (low pH) separations in four consecutive injections (experiments). In each experiment, a five-step fractionation using 10.8, 12.4, 15.4, 18.6, and 50% Eluent B (100% ACN) was performed in the first dimension. T43 ENL peptide is eluted only in Fraction 4, demonstrating great reproducibility of the first dimension fractionation. In addition, the retention time reproducibility (0.05% RSD) for the same peptide over 48 hours of separation demonstrates the stability of the 2D-UPLC system.
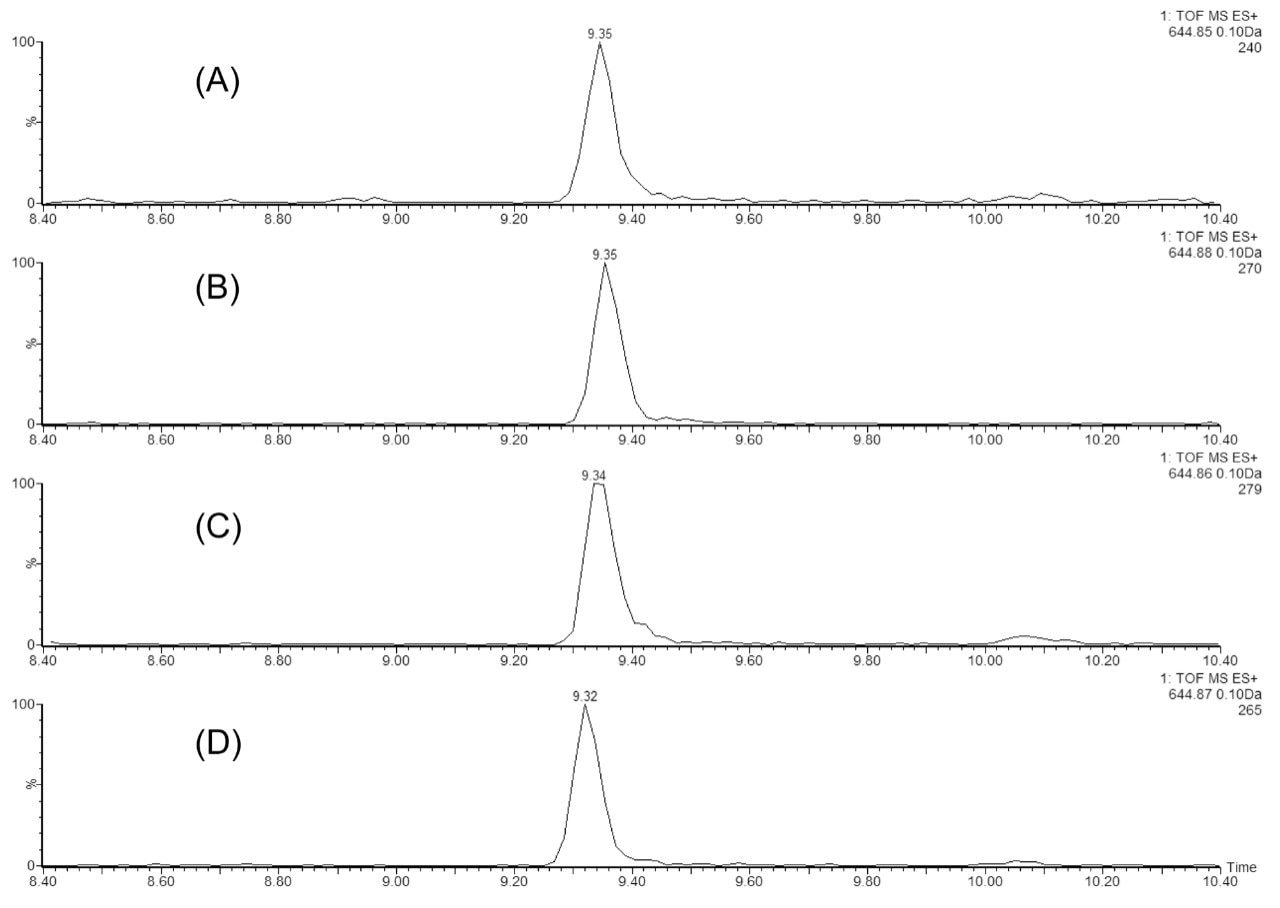
In the second experiment, we investigated the ability of the 2D-UPLC system to maintain good chromatographic performance, independent of the number of fractionation steps in the first dimension. Figure 3 displays the extracted mass chromatograms of T43 peptide generated from the second dimension separations under four fractionation schemes: “simulated 1D” (single step elution from 10.8 to 50% of Eluent B), 3-step, 5-step, and 10-step fractionations, respectively. The system demonstrates the highly reproducible retention time over the different operation schemes (0.15% RSD). The 10-step fractionation experiment revealed an important aspect regarding the high pH/low pH 2D separation of peptides: peptides can elute entirely within a single fraction (5/10) even with a relatively narrow elution step-gradient (1.9% B). As shown in Figure 4, no T43 peptide could be detected in the previous or the following fractions.
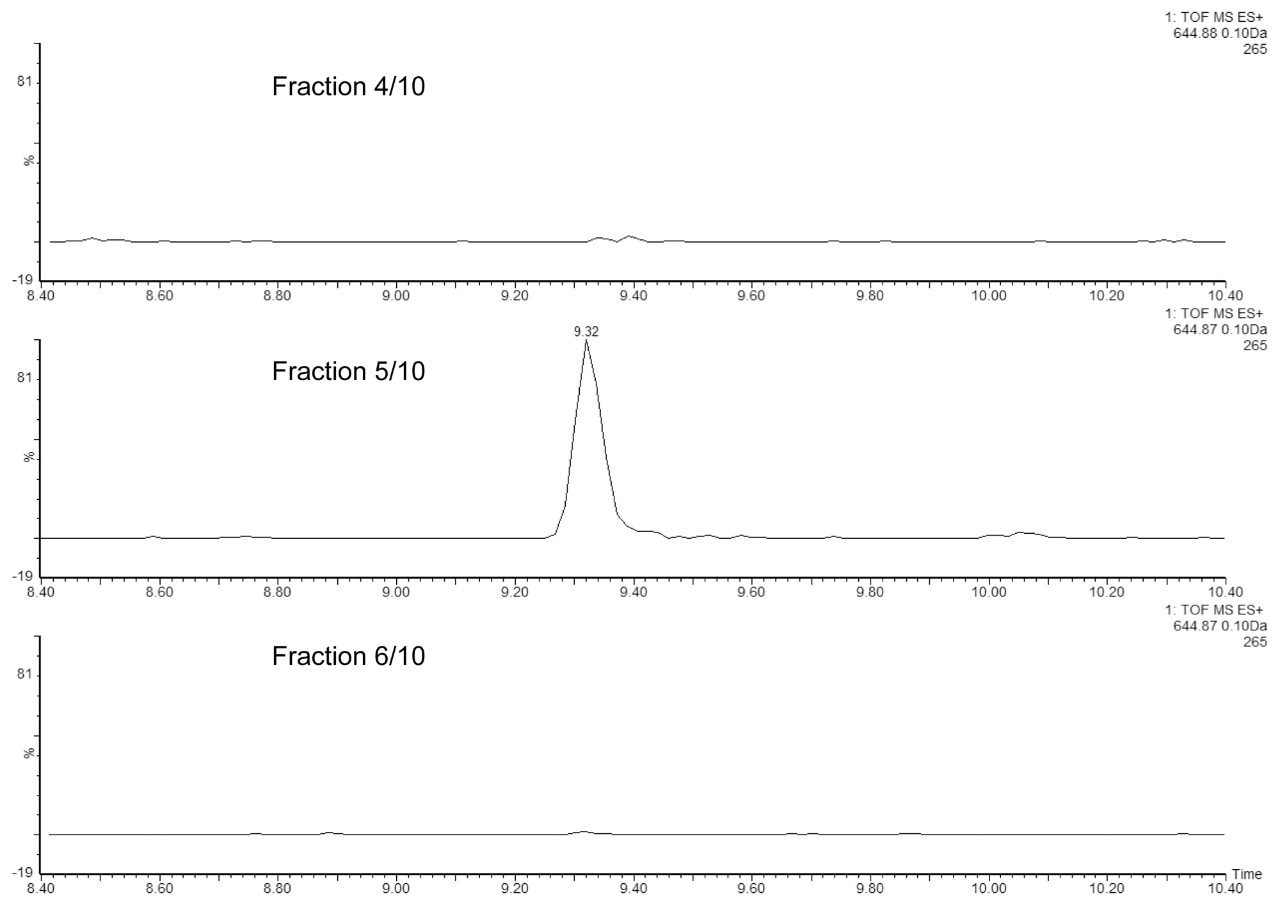
The chromatographic performance of the 2D-UPLC setup can be maintained, regardless of the number of fractionation steps. The reproducibility of the 2D-UPLC setup with respect to the retention time, peak width/shape, and the intensity of the ESI-MS signal within four consecutive experiments demonstrates the absolute trust a laboratory studying HCPs can place in the system. Overall, the data demonstrates the ability of the high pH XBridge Column to retain peptides over the duration of the fractionation experiment (six to 10 hours) without measurable sample losses.
To test the analytical capability of the 2D-UPLC/MSE technology for identification of low-abundance HCPs in biopharmaceuticals, we analyzed six mAb (PTG1) samples that were expressed by DG-44 (samples labeled A1, A2, B1, and B2) and CHO-S hamster cell lines (samples C and D).
Each of the samples was purified by two different protein-A chromatography columns. This single-step purification protocol was not designed to achieve fully optimized purification for the highest purity of mAb, rather it provided good test samples to examine the capability of the LC-MS assay to identify HCPs from relatively complex biological samples.
A relatively large number of CHO proteins (about 40) were co-purified (see Table 1) with the mAb target. Five proteins standards (originating from other species than the host hamster cells) were spiked in the PTG1 preparations before tryptic digestion. These protein standards serve as an internal control to probe the dynamic range of the assay and to provide internal references for quantification of HCPs using the summed signal of the three best responding peptides of each protein identified in the analysis.8
As shown in Table 1, a total of 37 HCPs across six PTG1 preparations were identified. Because the CHO protein database is not available in the public domain, these HCP proteins were identified using the mouse/hamster homology search. The measured HCP concentrations varied widely, from 10 to 3000 ppm. Four out of five spiked protein standards (except the lowest abundance ENL) were identified in all samples (two out of three replicates). Most of the HCPs identified in this study were high-abundance CHO proteins.
Table 1 indicates that the HCP composition/concentration significantly depends on the cell lines used for PTG1 expression. Total HCP concentrations for samples C and D produced by the CHO-S cell line are significantly lower than the total HCP concentrations measured for DG-44 (samples A1, A2, B1, and B2). In addition, the protocols used for Protein A purification of PTG1 also influences the HCP composition/concentration as suggested by Table 1.
Once a purification process is established, organizations need to monitor the known HCPs to prove that their process is well-controlled. They may also need to demonstrate to the regulator that the claims made for the product are consistent throughout a number of batches.
UPLC/MRM-MS methodology can provide this information in a rigorous and objective manner that does not rely on operator interpretation. Furthermore, changes in process methodology may also mean that new ELISAs may take months to develop, whereas UPLC/MRM-MS methods can be changed within minutes to accommodate new proteins.
Therefore, a 20-min UPLC/MRM-MS method was developed on the basis of the discovery results from the 2D-UPLC-MSE step. The UPLC/MRM-MS assay is developed to rapidly monitor the concentration changes of the previously identified HCPs prepared from a variety of experimental conditions. Twenty HCPs were selected from the list of identified proteins in the six samples.
In total, 58 transitions for 29 peptides representing the twenty HCPs were monitored in MRM experiments. The assay generated highly reproducible measurements with an average peak area RSD of 13.8% for the entire MRM dataset. The results demonstrate that UPLC/MRM-MS methodology offers an efficient method for high-throughput HCP monitoring during the late stage of biopharmaceutical purification. In addition, an MRM assay provides an easy method for absolute quantification of each individual HCP by using spiked-in isotopically labeled peptides.
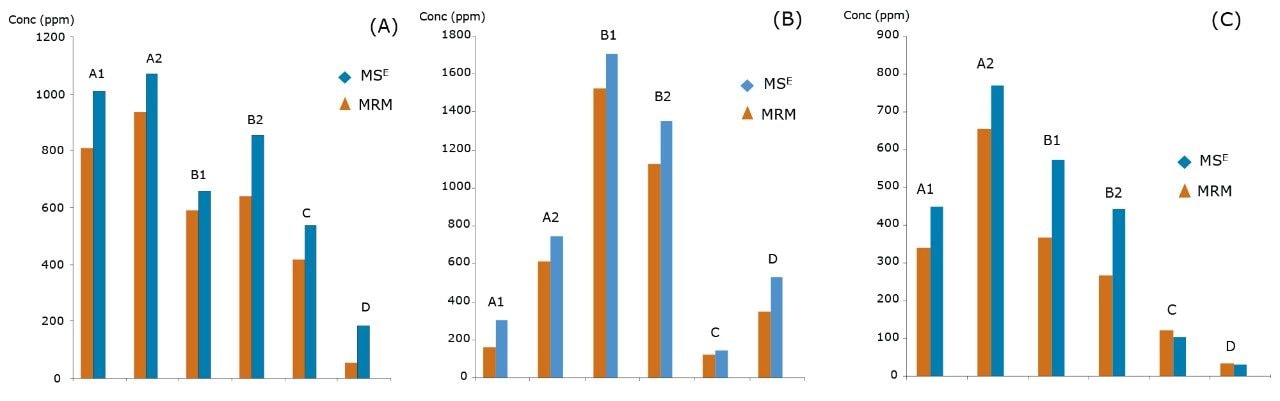
It is also important to understand the correlation between the different UPLC-MS techniques used. Three HCPs (out of 20 HCPs monitored in MRM experiment) were quantified using spiked-in isotopically labeled peptides. This method matches the technique used in the industry today to obtain absolute quantification values. The MRM results correlate well with the MSE quantification across all six samples, as shown in Figure 5A–C.
720004043, July 2011