
For forensic toxicology use only.
This is an Application Brief and does not contain a detailed Experimental section.
This application brief demonstrates the creation, contents and use of a compound database for targeted TOF/MS based screening using Pos±tive Software.
Targeted screening is the analysis of a sample for the presence or absence of known compounds. It is usually semi-quantitative, using standard solutions of commonly encountered compounds to give the user an approximation of concentration within the sample. This allows the analyst to identify if their compounds of interest are present in quantities that will be close to, or exceed relevant legislative levels. This procedure is increasingly performed using a time-of-flight (TOF) instrument, such as Xevo QTof MS, because they enable many hundreds of compounds to be screened in a single run, and provide a comprehensive historical record of sample components, allowing post-acquisition processing and targeting of compounds of interest. Positive results will generally be re-analyzed using a complimentary technique such as tandem quadrupole MS/MS.
One of the key tools required for a successful targeted screen is information regarding the compounds of interest. This is stored as a database and used by the processing software to detect and identify the compounds of interest.
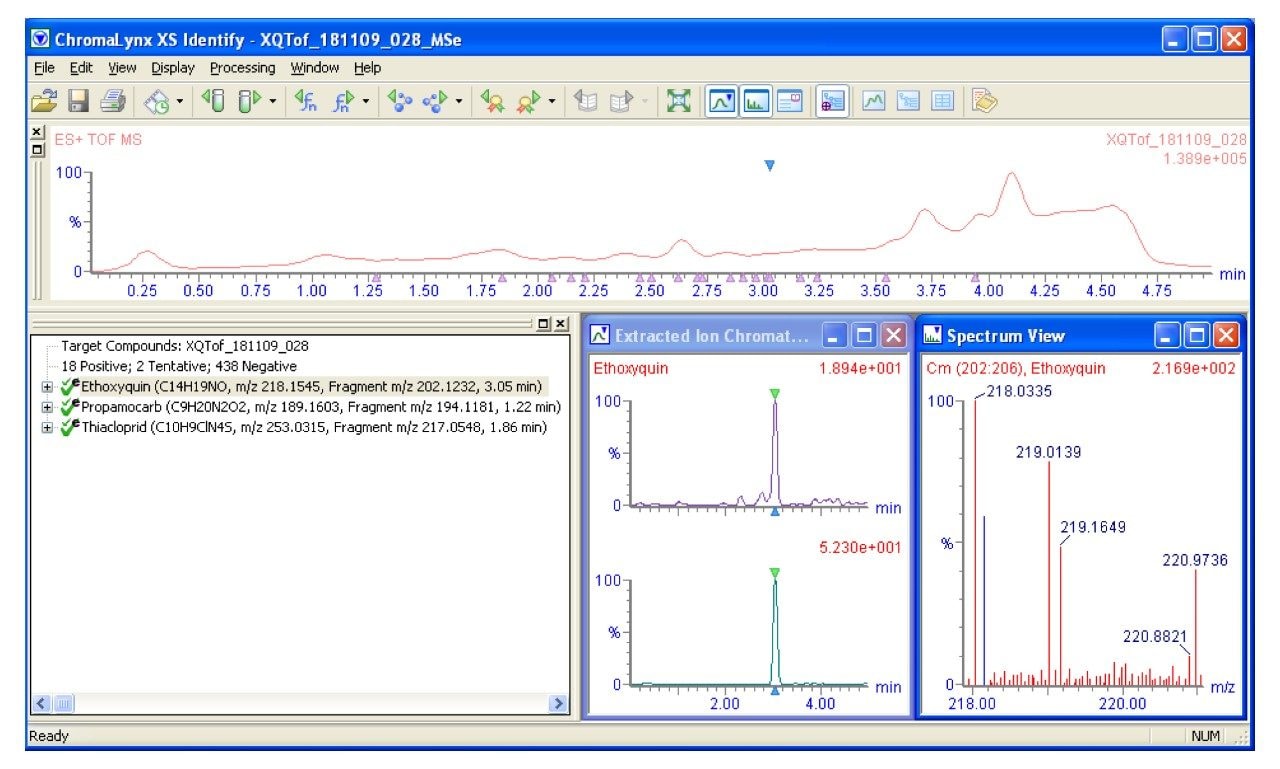
The chemical analysis database is a list of compounds for which the exact mass and retention time data have been collected using a specified LC gradient. This includes 450 pesticides and 150 veterinary drugs, as well as the [M+H]+ ion, [M+Na]+, [M+K]+, [M+NH4]+, and [M-H]- ions that have all been assessed. The database also includes fragment ion data which can be used to improve confidence in identification of compounds when running with MSE enabled. MSE is a patented data independent acquisition technique that provides a simple, unbiased, parallel route to delivering exact mass molecular (MS) and product ion (MSE) information from every detectable component, without the need for multiple injections. Figure 1 shows part of the database.

Solvent standards were run for each compound currently in the database. Individual standards make it easier to identify the correct peak for each compound and should be used if possible. Mixtures or matrix based samples can be used but this may add extra complexity to the interpretation of the result. Each compound was analyzed in both positive and negative ion mode with MSE enabled.
After the solvent standard was run the chromatogram was assessed to see which of the ions or adduct ions mentioned earlier were present. The data from the strongest signal was collated and entered into the database. The spectrum from this peak was also assessed to identify fragment ions. Fragment ions with the greatest abundance were noted and entered.
The system should be calibrated and equilibrated with the supplied method parameters so that the retention time data is matched to that in the database. If substitutions are made to the method that involve different parameters, such as column or mobile phase, then the retention times are likely to vary. It is important to use the specified conditions as Posi±ive Software will greatly reduce the amount of time for processing and data review with the correct retention times. Full details of this method can be found in the appendix at the end of the document.
Posi±ive data processing software can be utilized to easily identify which compounds are present above a user-defined threshold level in the sample (e.g. a legislative limit), identifying them as presumptive positive for confirmation using a validated LC-MS/MS method. The database is easily loaded into the processing method. The processing software takes the information from the database and seeks out exact mass – retention time pair matches using ChromaLynx XS Application Manager. If these are present, then it promotes this data through to TargetLynx Application manager where a semi-quantitative result is produced. User-defined values set the boundaries for acceptance criteria. A calibration reference compound can be specified for every compound in the database. This means that all the compounds being screened do not need to have an associated calibration line, reducing costs of expensive analytical standards.
Finally, a report can be produced from Posi±ive Software that shows which samples have compounds present above user-defined values. This is different from historical reports as the large numbers of negative results are removed, reducing the time for data analysis after the samples have been run.
Previously, the exact mass and the retention time have been used to positively identify the compounds present. Improved confidence could be obtained with a second injection where fragmentation occurs and spectral information is used to confirm identity. The unique MSE acquisition mode of Waters Xevo QTof MS simultaneously captures data for low and high collision energy which is stored by MassLynx Software in two different channels. When data processing, these two channels are analyzed and the spectrum of the fragmented parent is used for verification, giving the extra layer of identification with no extra work for the analyst. Figure 2 shows a screenshot of a processed sample using Posi±ive Software. The identity of the compound in question has been proved using MSE. The lower green trace shows the parent ion from acquisition channel 1, and the upper purple trace shows a structural fragment in acquisition channel 2.
This process allows the user to screen for the parent ion of compounds whilst collecting comprehensive fragment information. The benefit of this approach is that increased confidence in compound identity can be achieved in just one injection, reducing the amount of false positive results and removing the need for another identifying injection. This approach also works when the retention time is not known.
As more samples are run, the database has the ability to increase in size with the addition of more compounds. Further versions of this database will be created and distributed when demand is high. Possible additions include different compound classes and the latest pesticides in the marketplace. This database can be easily modified in house to accommodate individual needs with the addition of user compounds.
A well characterized database containing exact masses and retention times for compounds is required for a successful targeted screening experiment to be performed.
Here we have described the creation and use of such a database with Waters Posi±ive Software, where the user-editable database is easily loaded into a data processing method.
This database adds a further dimension of quality to analyst’s results as confidence of identity is aided with the use of fragment ion data through MSE.
The use of Posi±ive Software greatly reduces the amount of time an analyst spends reviewing results as its report does not need to contain long list of samples showing a negative result.
Weigh 7.7 g of ammonium acetate and dissolve using 100 mL water, which yields a 1 M solution.
|
Mobile phase A: |
10 mL of 1 M ammonium acetate and 990 mL water |
|
Mobile phase B: |
10 mL of 1 M ammonium acetate and 990 mL methanol |
|
Weak wash solvent: |
Water |
|
Strong wash solvent: |
Methanol |
|
Seal wash: |
90:10 water: methanol (v/v) |
|
Lock mass compound: |
Leucine enkephalin at 500 pg/μL |
Suitable for both positive and negative ion modes, prepare a stock 1 mg/mL solution of leucine enkephalin solution in water. Take 500 μL of 1 mg/mL solution and add to 500 mL acetonitrile and 500 mL water and 1 mL formic acid. If it is necessary to prepare a more concentrated solution to achieve the required signal, simply add more stock solution.
|
Column: |
ACQUITY UPLC BEH C18, 2.1 x 50 mm, 1.7 μm, Part no. 186002350 |
|
Column temp: |
45 °C |
Purge column after use with water/ methanol gradient to remove any buffer to help column lifetime.
|
Electrospray, positive, and negative ion modes. |
|
|
Capillary (kV): |
1.0 |
|
Sampling cone: |
30.0 |
|
Extraction cone: |
4.0 |
|
Source temp: |
120 °C |
|
Desolvation temp: |
550 °C |
|
Cone gas flow: |
50 L/Hr |
|
Desolvation gas flow: |
1000 L/Hr |
|
Purge gas flow: |
1000 L/Hr |
|
Collision energy: |
6.0 |
|
Scan time: |
0.2 s |
|
MSE parameters |
|
|---|---|
|
Low energy: |
6 V |
|
High energy ramp: |
25 - 35 V |
|
Time (min) |
Flow rate |
%A |
%B |
Curve |
|---|---|---|---|---|
|
Initial |
0.600 |
98.0 |
2.0 |
— |
|
0.10 |
0.600 |
98.0 |
2.0 |
6 |
|
3.75 |
0.600 |
1.0 |
99.0 |
6 |
|
4.25 |
0.600 |
1.0 |
99.0 |
6 |
|
4.26 |
0.600 |
98.0 |
2.0 |
11 |
|
Total run time: |
5.00 min |
|||
|
Injection volume: |
10.00 μL full loop, 20 μL loop with PLNO can also be used. |
Tubing on all instrumentation was cut to a minimum. A standard difference (offset) in retention times is often due to differences in this tubing between instruments.
All data collected for this database must be acquired using this UPLC gradient with the parameters specified in this package. MSE data should be acquired to attain fragment ion data. A high solvent standard was injected to make peak identification easy.
720003341, February 2010