A Rapid Approach to Metabolite Identification Using Xevo MRT Mass Spectrometer and MassMetaSite Software
Richard Lock, Robert Plumb, Lee Gethings
Waters Corporation, United States
Published on November 25, 2025
Abstract
Metabolite identification is a critical component of drug discovery and development, offering essential insights for lead compound selection, safety assessment, interspecies comparisons, and clinical support. In the early stages of drug discovery, where numerous new chemical entities require evaluation, a rapid and efficient metabolite identification solution is vital.
This application note presents a high-throughput platform for metabolite identification, integrating the Waters Xevo™ MRT Mass Spectrometer with the ACQUITY™ Premier UPLC™ Chromatography System and MassMetaSite software (Mass Analytica). The platform was used to analyze urinary metabolites of methapyrilene, a 2-thiophene-based H1-receptor antagonist, following oral administration to male rats. The analysis was completed using a rapid 5-minute LC-MS method.
A total of 42 metabolites were identified, each with MS1 and MS2 mass errors below 1 ppm. The direct data transfer from waters_connect™ MS Software to MassMetaSite software streamlined the workflow, simplifying the analysis and enhancing throughput.
Benefits
- The Xevo MRT Mass Spectrometer delivers sub-ppm mass accuracy in both MS1 and MS2 modes, enabling precise localization of biotransformation sites
- The fast DIA and DDA acquisition capabilities of the Xevo MRT Mass Spectrometer make it ideal for high-resolution mass spectrometry (HRMS), supporting short LC run times and significantly boosting throughput
- Seamless data transfer from waters_connect Software to Mass Analytica’s MassMetaSite metabolite identification software simplifies the workflow, efficiently converting LC-MS data into clear metabolite identifications
Introduction
Metabolite identification is a vital step in the pharmaceutical R&D process. It provides essential insights during drug discovery supporting in vitro screening, lead candidate selection, and pharmacokinetic profiling, and continues to play a key role in development by informing interspecies comparisons, dose escalation safety studies, and clinical evaluations.
In the discovery phase, the volume of new chemical entities is high, necessitating rapid, broadly applicable methodologies that do not require compound-specific customization. At this stage, limited information is available about a compound’s metabolic fate. As such, identification relies on mass spectral characteristics and known biotransformation pathways.
In contrast, the development phase demands comprehensive metabolite profiling across species, though throughput is less critical due to fewer compounds. By this point, prior metabolic data (e.g., from in vitro studies) often exists, allowing researchers to confirm known metabolites and identify new ones. Additionally, radiolabeled compounds may be used to highlight drug-related species.
To meet these needs, a high-throughput, accurate, and reliable system for metabolite detection, data mining, and identification is essential. This application note describes a solution combining the Waters Xevo MRT Mass Spectrometer, ACQUITY Premier UPLC Chromatography System, and MassMetaSite software. This integrated platform leverages fast chromatographic performance, high-speed MS1 and MS2 data acquisition with sub-ppm accuracy, and intelligent data analysis to rapidly detect and assign drug metabolites.
Experimental
Animal Study
Rat urine samples were obtained following the repeat oral (PO) administration of methapyrilene (MP) to male Wistar rats (n=18) at 0, 50, or 150 mg/Kg. Urine was collected pre-dose and 16 hours following dosing on day 3 and 5 of the study. The animal study was performed by Evotec SAS (Toulouse, France), following full management and ethical review according to national and EU guidelines. Full details of this study are given in publication by Wilson et-al.1
Sample Preparation
Urine samples were prepared by mixing 10 µL urine with 40 µL ACN:MeOH (90:10 v/v). Samples were then vortex mixed prior to storing at -20 °C for 2 hours. Following incubation, samples were vortex mixed and centrifuged at 25,000 g for 5 minutes. The resulting supernatant was then diluted 1:5 (v/v) with distilled water and transferred to LC-MS total recovery vials (p/n: 186002805).
LC-MS Data Analysis
The samples were analyzed by reversed–phase chromatography using an ACQUITY Premier UPLC Chromatography System coupled to a Xevo MRT Mass Spectrometer. The chromatography was performed on a CORTECS™ C18, 1.6 µm, 2.1 x 50 mm analytical Column (p/n: 186007114). Following sample injection (1 µL), the column was eluted with a 5–95% organo – aqueous gradient over 5 minutes at 0.6 mL/min, where mobile phase A comprised 0.1% formic acid containing 1 mMol ammonium formate and mobile phase B 95:5 ACN:water (v/v), 0.1% formic acid, 1 mMol ammonium formate).
Analytes detection was performed in +ve ESI DIA mode using the conditions described in Table 1.
MS Conditions
|
Parameter: |
Value |
|
Ionization mode: |
ESI +ve |
|
Desolvation gas: |
800 mL/min |
|
Cone gas: |
50 mL/min |
|
Capillary voltage: |
1 kV |
|
Desolvation temperature: |
600 °C |
|
Source temperature: |
120 °C |
|
Collision energy: |
Ramp from 5–50 eV |
|
Cone voltage: |
30 V |
Data Management
|
Chromatography software: |
waters_connect Software Platform |
|
MS software: |
waters_connect Software Platform |
|
Informatics: |
MassMetaSite (Mass Analytica, Sant Cugat del Vallés, Spain) |
Methapyrilene (MP), formally known as N,N-dimethyl-N'-pyridin-2-yl-N'-(thiophen-2-ylmethyl)ethane-1,2-diamine, is a H1-receptor antagonist that was previously marketed as a sedative and antihistamine. However, it was withdrawn due to its carcinogenic effects in rats following chronic administration. These toxic effects have been linked to chemically reactive metabolites formed through bioactivation of the thiophene ring.1,3 Additional metabolic pathways, including demethylation, oxidation, and conjugation, have also been reported.4
This study aimed to investigate the metabolism of MP in rats after repeated oral dosing. Rat urine samples were analyzed using the Xevo MRT Mass Spectrometer in DIA mode. The resulting LC-MS data underwent peak picking, background subtraction, and were processed using MassMetaSite software.
To facilitate analysis, LC-MS data were exported to MassMetaSite via the data conversion feature in waters_connect Software Platform, generating .mzML files. Integration with the waters_connect Hub API enabled streamlined data transfer, as illustrated in Figure 1.
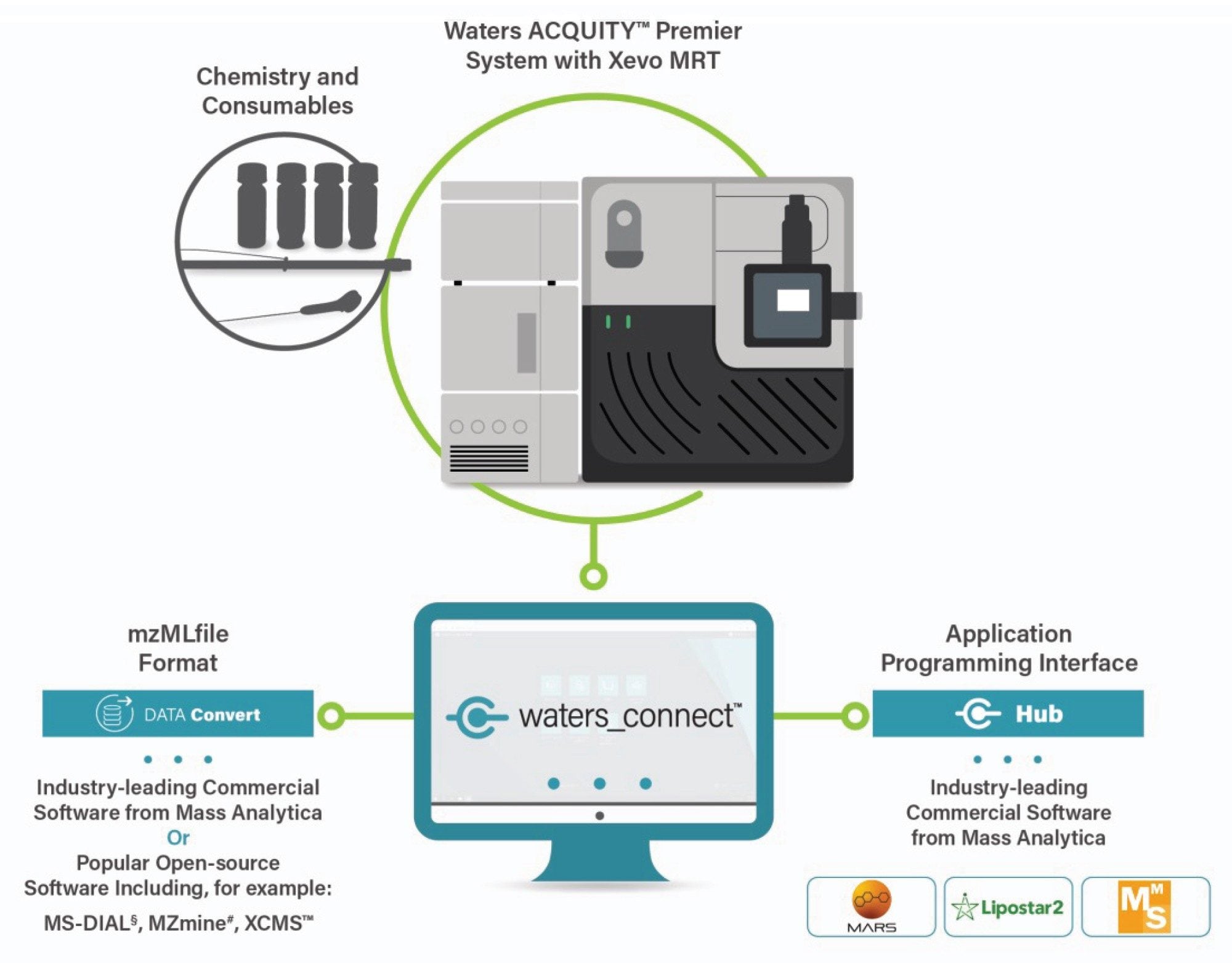
Results and Discussion
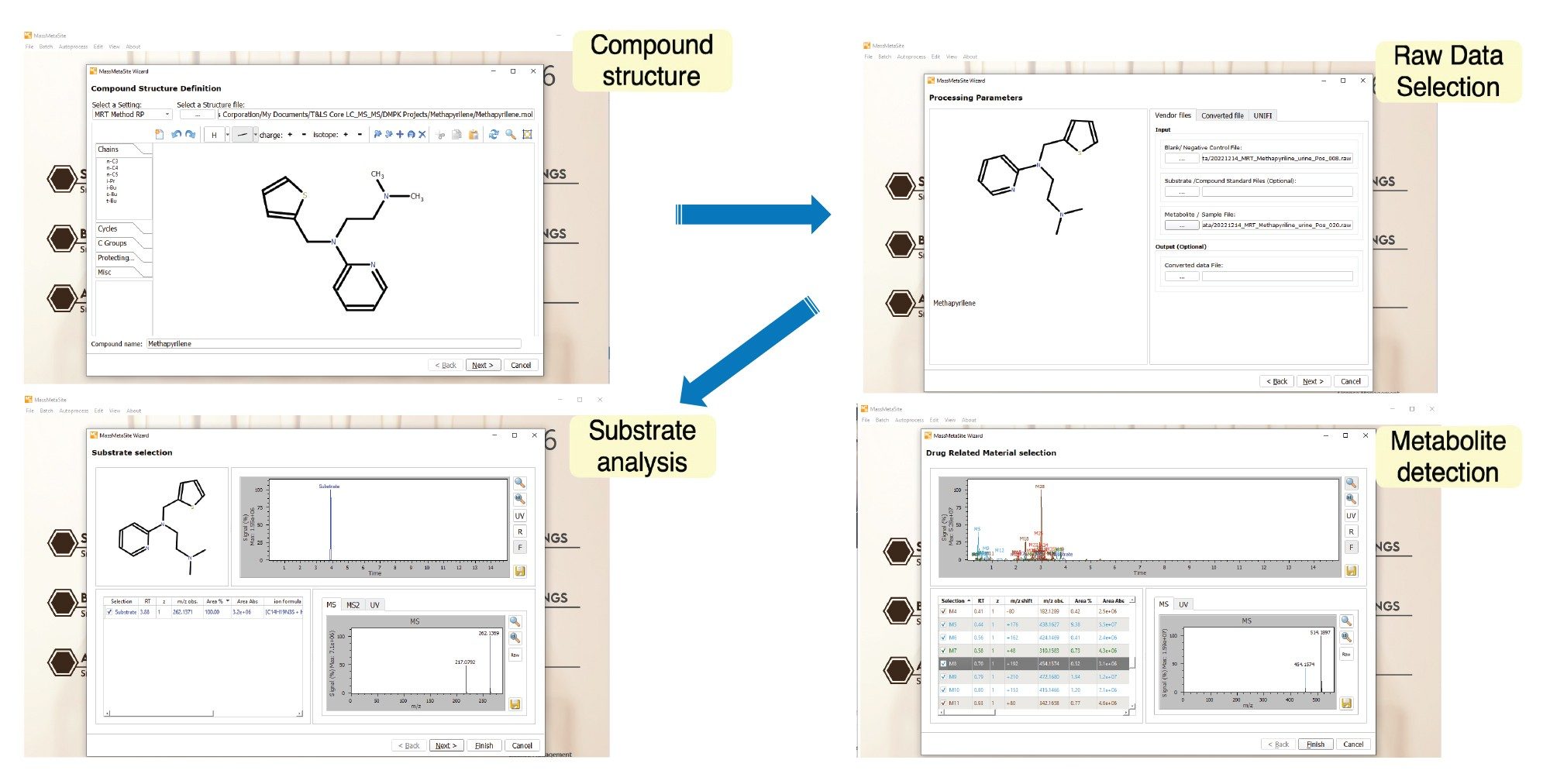
Metabolite identification using MassMetaSite follows a straightforward, step-by-step workflow:
- Create an MS processing method – This defines key parameters such as MS polarity, instrument type, acceptable mass error range, and the biotransformations to include in the analysis.
- Input the drug structure – The chemical structure of the compound under investigation is entered into the software.
- Select LC-MS data files – This includes files for the drug-treated sample, a blank matrix, and the drug substrate.
Once these steps are completed, the software automatically detects the substrate peak and extracts both precursor (MS1) and product (MS2) ion data. Using this information, along with common biotransformation rules, MassMetaSite searches for potential drug-related peaks. The MS2 data and molecular structure are then used to pinpoint the site of metabolism.
The results are displayed in an interactive browser for further review and analysis (see Figure 2).
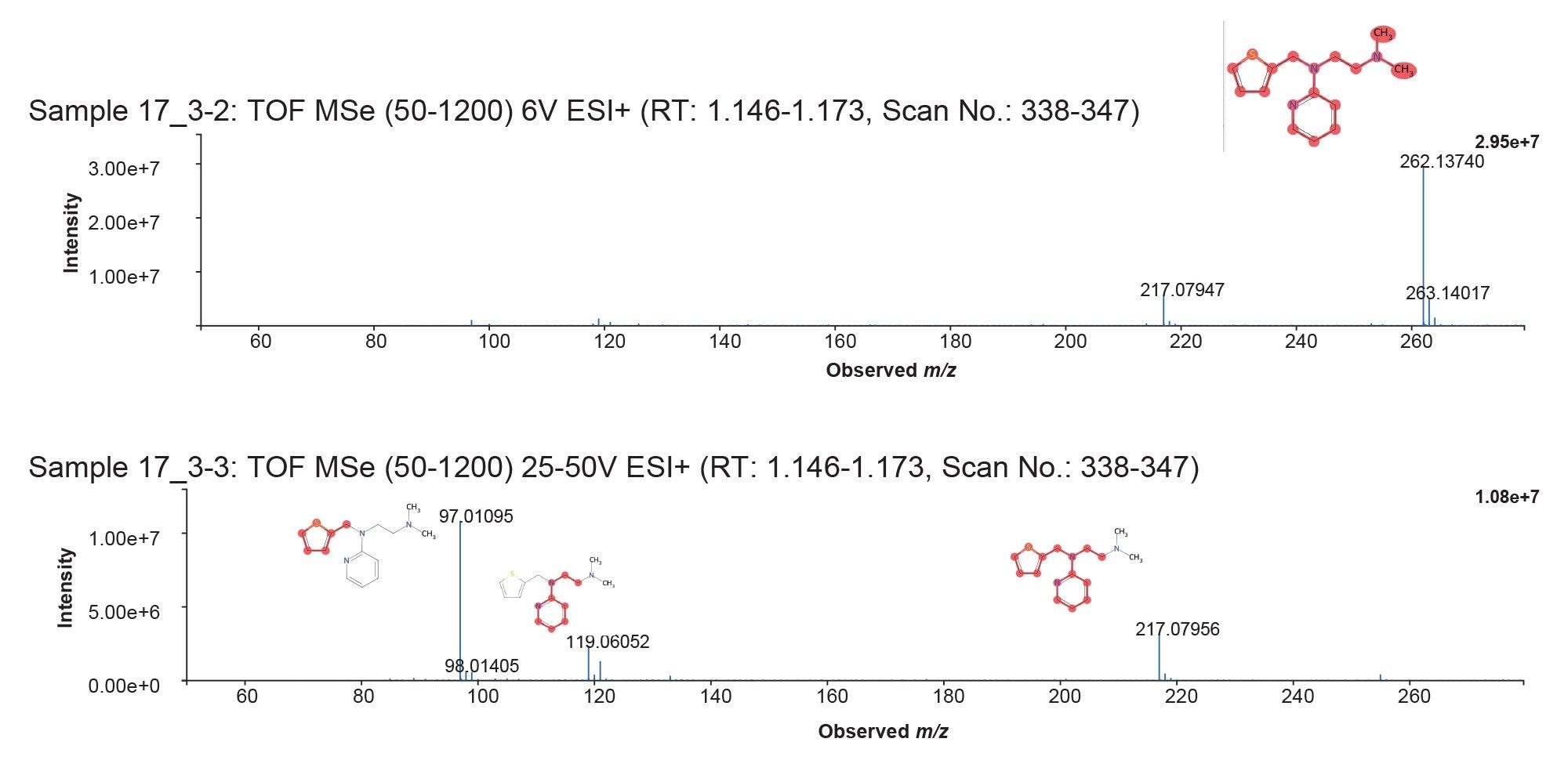
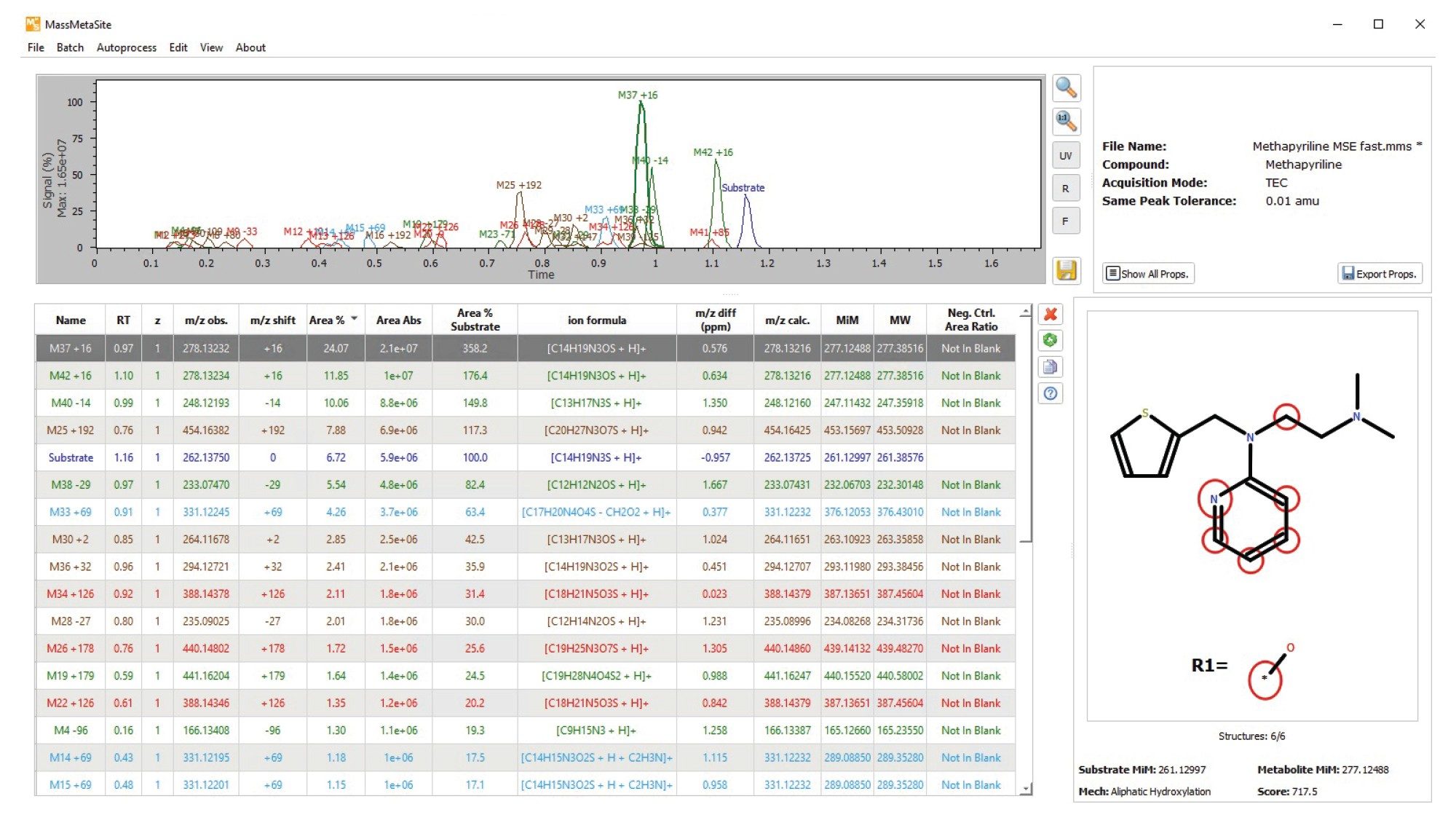
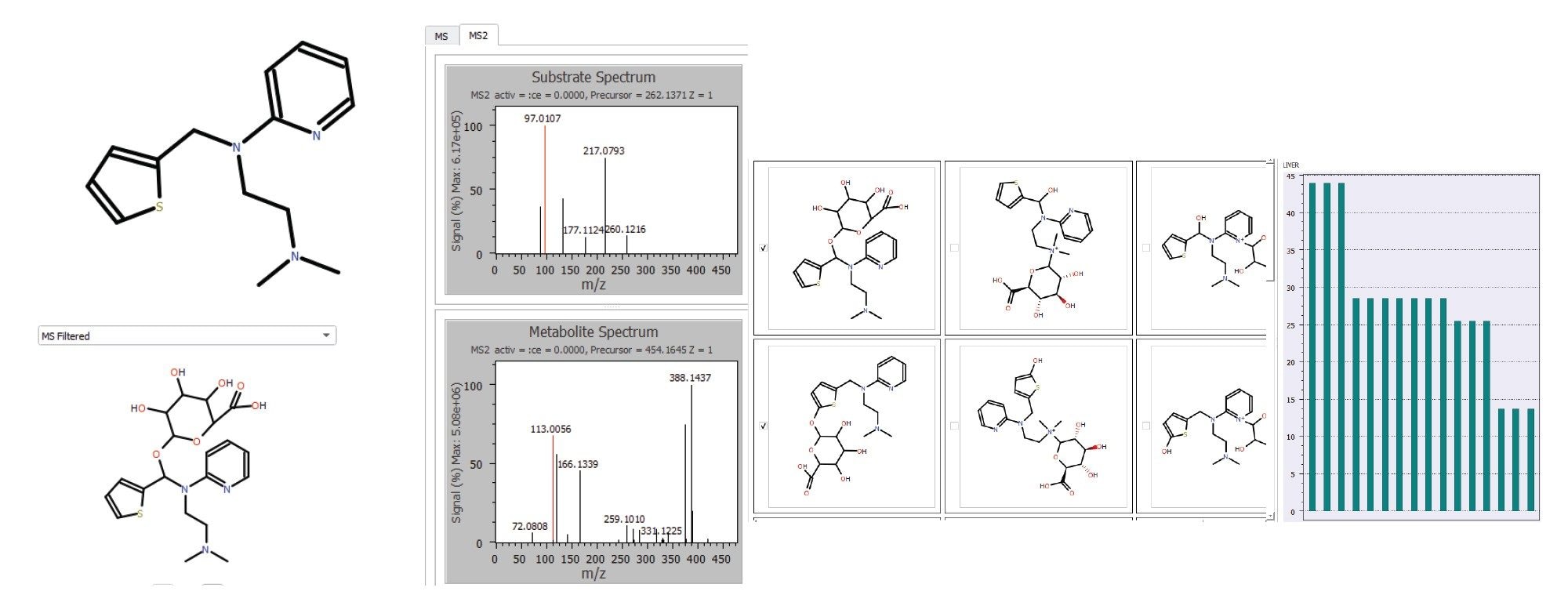
Review of the LC-HRMS data revealed that methapyrilene (MP) eluted at a retention time of 1.16 minutes, with a base peak at m/z 262.13750 and a mass error of –0.957 ppm (Figure 3). In total, 42 drug-related metabolites were detected, all with mass errors below 1.7 ppm— most of them under 1 ppm. These results are summarized in the MassMetaSite browser (Figure 4).
The earliest metabolite was observed at 0.12 minutes, while the latest drug-related peak appeared at 1.10 minutes. Despite the narrow chromatographic peak widths (averaging just 3 seconds at the base), the Xevo MRT System successfully acquired more than 10 spectra in both MS1 and MS2 channels. This high spectral density enabled precise definition of each LC peak.
Metabolite data can be filtered by mass error, retention time, m/z value, or percentage area. Each metabolite entry can be individually reviewed, allowing direct comparison of its MS1 and MS2 spectra with those of the parent drug.
MassMetaSite software also proposes a likely structure for each metabolite, along with alternative possibilities. These are presented with a probability plot that illustrates the plausibility of each structure. For example, the metabolite with m/z 454.16382 (shown in Figure 5) is predicted to be an O-glucuronide attached to the thiophene ring.
The ability of the Xevo MRT System to acquire MS1 and MS2 data with sub-ppm mass accuracy greatly enhances the software’s capability to pinpoint the site of biotransformation. Overall, the data processing time was just a few minutes, significantly improving both throughput and confidence in the results.
The concentration of individual drug metabolites in plasma can vary significantly. Some biotransformation pathways are more favorable, resulting in major metabolites that dominate the route of elimination, while others produce minor metabolites present only at low concentrations. Detecting and identifying both major and trace metabolites is crucial, as even low-abundance species may be toxic or pharmacologically active.
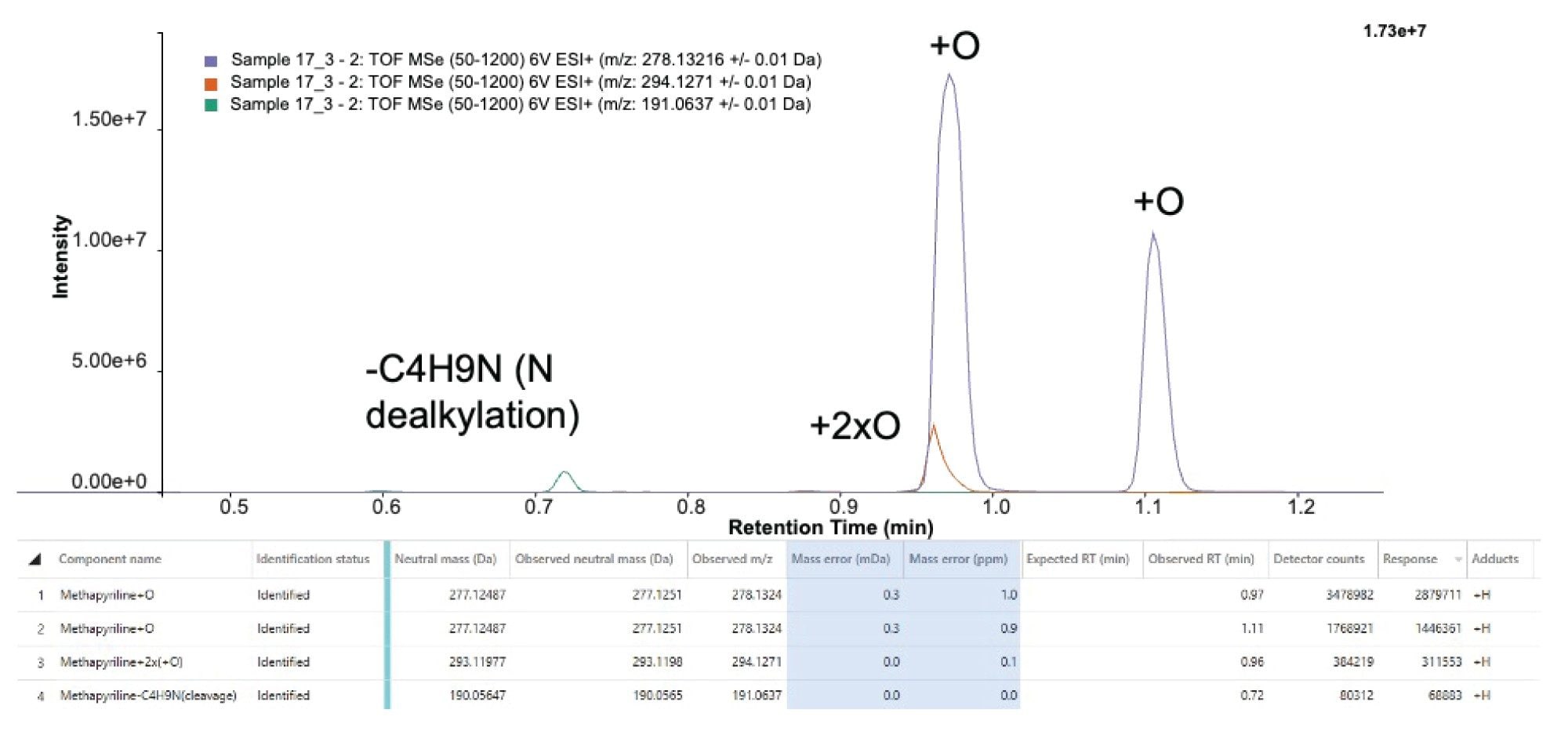
Methapyrilene is a prime example of a drug with complex metabolism. Previous studies (Wilson et al.1) have identified over 100 minor drug-related species. To capture this full metabolic profile, the mass spectrometer must offer a wide linear dynamic range.
The Xevo MRT Mass Spectrometer meets this need with its innovative dual-gain ADC GPU combination, delivering a dynamic range exceeding five orders of magnitude. Figure 6 demonstrates this capability, showing consistent sub-1 ppm mass accuracy across metabolites with varying intensities—including N-dealkylated (tR = 0.72 minute), dihydroxyl (tR = 0.96 minute), oxidation (tR = 0.97 minute), and N-oxide (tR = 1.11 minutes) forms of methapyrilene.
Conclusion
Rapid and accurate metabolite identification is essential to support pharmaceutical research and development. With the growing number of compounds requiring DMPK (drug metabolism and pharmacokinetics) assessment, there is a clear need for a high-throughput platform capable of both analyzing samples and efficiently processing the resulting data.
This application note describes a robust solution that combines:
- Fast, high-resolution chromatography from the ACQUITY Premier UPLC System
- Sub-1 ppm mass accuracy and wide dynamic range from the Xevo MRT Mass Spectrometer
- With the advanced data processing capabilities of MassMetaSite software
Using a rapid 5-minute generic reversed-phase LC gradient, this platform successfully detected and identified over 40 in vivo metabolites of methapyrilene. The exceptional mass accuracy of the Xevo MRT Mass Spectrometer in both MS1 and MS2 modes significantly enhances confidence in the proposed metabolite structures.
References
- Plumb RS, Lai SK, Gethings LA, Trengove R, Hancock P, Wilson ID. An investigation of the plasma and urinary metabolite profiles of the hepatotoxin methapyrilene in the male Wistar rat. J Pharm Biomed Anal. 2025 15;264:116976. doi: 10.1016/j.jpba.2025.116976.
- M.E. Graichen, D.A. Neptun, J.G. Dent, J.A. Popp, T.B. Leonard. Effects of methapyrilene on rat hepatic xenobiotic enzymes and liver morphology, Fundam. Appl. Toxicol. 5 (1985) 165-174. doi: 10.1016/0272-0590(85)90061-2.
- G.S. Ratra, C. Cottrell, C.J. Powell. Effects of induction and inhibition of cytochromes P450 on the hepatotoxicity of methapyrilene, Toxicol. Sci. 46 (1998) 185–196. doi: 10.1006/toxs.1998.2513.
- G.S. Ratra, C.J. Powell, B.K. Park, J.L. Maggs, S. Cottrell. Methapyrilene hepatotoxicity is associated with increased hepatic glutathione, the formation of glucuronide conjugates, and enterohepatic recirculation. Chemico-Biological Interact. 129 (2000) 279–295. doi: 10.1016/s0009-2797(00)00253-2.
720009145, November 2025