For research use only. Not for use in diagnostic procedures.
In this application note, we present a metabolomics study that combines targeted and untargeted approaches for breast cancer biomarkers analysis.
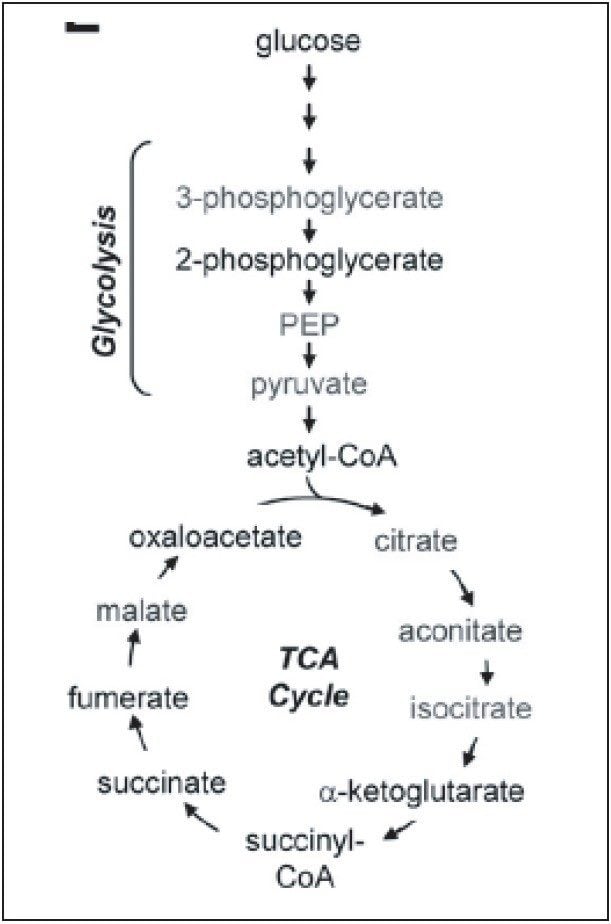
Breast cancer is one of the top five cancers that affect human lives seriously. Therefore, it is of great importance to discover the best ways to study this disease. Metabolic reprogramming is required both during the initial breast cancer transformation process (primary tumor) and during the acquisition of metastatic potential (metastases), shown in Figure 1. The reprogramming process includes altered flux through glycolysis and the pentose phosphate pathway (PPP), resulting in increased fatty acid synthesis needed for proliferation.
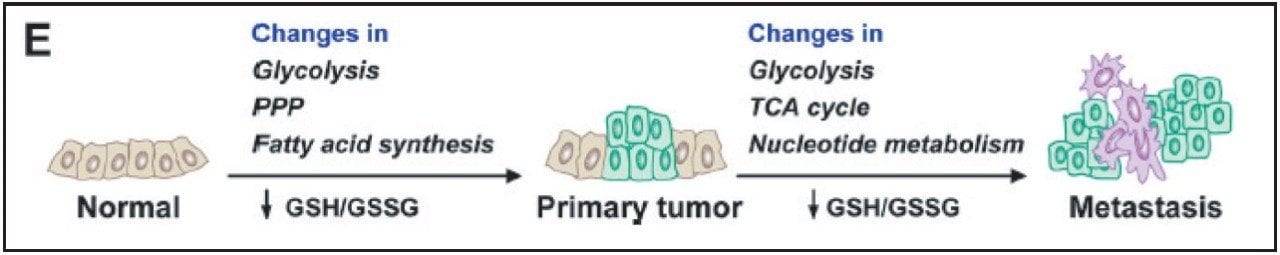
Reactive oxygen species produced during tumor progression result in a decreased glutathione GSH (reduced)/GSSG (oxidized) redox pool, which impairs genome stability, tumor suppressor gene function, and control over cell proliferation. Continued GSH/GSSG depletion in the primary tumor may also contribute to general metastatic ability, and includes further changes in glycolysis and tricarboxylic acid cycle (TCA cycle) and increased nucleotide (PPP) flux for replication.
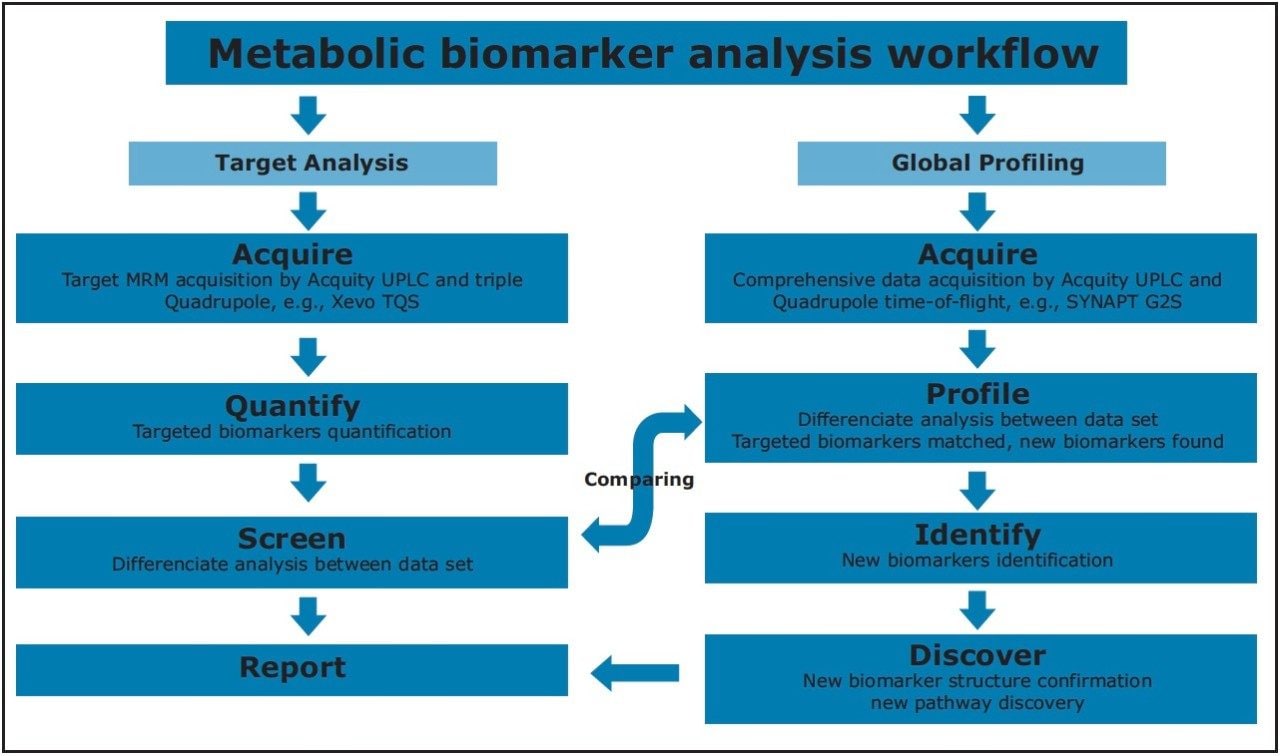
Staging of the metabolic reprogramming using metabolomics could pinpoint the metabolic processes that are essential for breast cancer transformation and invasiveness, which may yield biomarkers and new directions for therapeutics. In this application note, we present a metabolomics study that combines targeted and untargeted approaches for breast cancer biomarkers analysis. Figure 2 illustrates the workflow for this study.
Two rodent breast cancer cell lines, MTln3 (highly metastatic) and MTC (poorly metastatic), were used and cultured in Eagle’s minimal essential medium and supplemented with 5% fetal bovine serum (Invitrogen).
Cells were grown in 10-cm tissue culture dishes, and the media were replaced 24 h and 2 h prior to metabolite extraction. All samples were harvested at subconfluence.
Metabolism was quenched, and metabolites were extracted by aspiration of media and immediate addition of 4 mL of 80:20 methanol/water at 80 °C to simultaneously lyse cells and quench metabolism.
|
System: |
Waters ACQUITY UPLC System |
|
Column: |
ACQUITY UPLC HSS T3 Column 2.1 x 100 mm, 1.8 μm |
|
Column temp.: |
40 °C |
|
Flow rate: |
300 μL/min |
|
Mobile phase A: |
Water, 0.1% Formic Acid |
|
Mobile phase B: |
Acetonitrile, 0.1% Formic Acid |
|
Injection vol.: |
10 μL |
|
Time (min) |
%A |
%B |
Curve |
|---|---|---|---|
|
Initial |
99 |
1 |
Initial |
|
8.0 |
50 |
50 |
6 |
|
8.1 |
1 |
99 |
6 |
|
11.0 |
1 |
99 |
6 |
|
11.1 |
99 |
1 |
6 |
|
15.0 |
99 |
1 |
6 |
The SYNAPT G2 HDMS was operated in both positive and negative MSE modes. The capillary voltage used was 2.0 kV with the source and desolvation temperatures set at 120 °C and 400 °C, respectively.
In the MSE acquisition mode, the instrument alternates between a low and high collision energy state on alternate scans. This allows for collection of precursor and fragment ion information of all species in an analysis without the sampling bias that is introduced with other common methods, such as DDA where a specific m/z must be isolated before fragmentation.
The Xevo TQ MS was operated in both positive and negative MRM modes. The capillary voltage used was 2.0 kV with the source and desolvation temperatures set at 150 °C and 650 °C, respectively. The desolvation gas flow was set at 1200 L/hr and the collision gas (argon) flow 0.18 mL/min (4 x 10-3 mBar), with MS1/MS2 resolution at unit mass.
Targeted analysis was used to survey known metabolic pathways that are key to cancer aggressiveness, as outlined in Figure 1. Figure 4 shows many of the targeted metabolite markers that are elevated in the MTln3 cells from both the heat map (top) and relative change scale bar plot (bottom). Analysis of experimental data supports a Warburg effect cancer model.2 For highly aggressive MTln3 cells, high cytosolic NADH is indicated by a glycolytic/TCA cycle signature of an increased malate/aspartate shuttle, shown in Figure 6. High AMP levels for MTln3 cells suggest the elevated malate/aspartate shuttle cannot keep up with cellular energy needs. High levels of amino acids seen in MTln3 cells are necessary for cell growth.
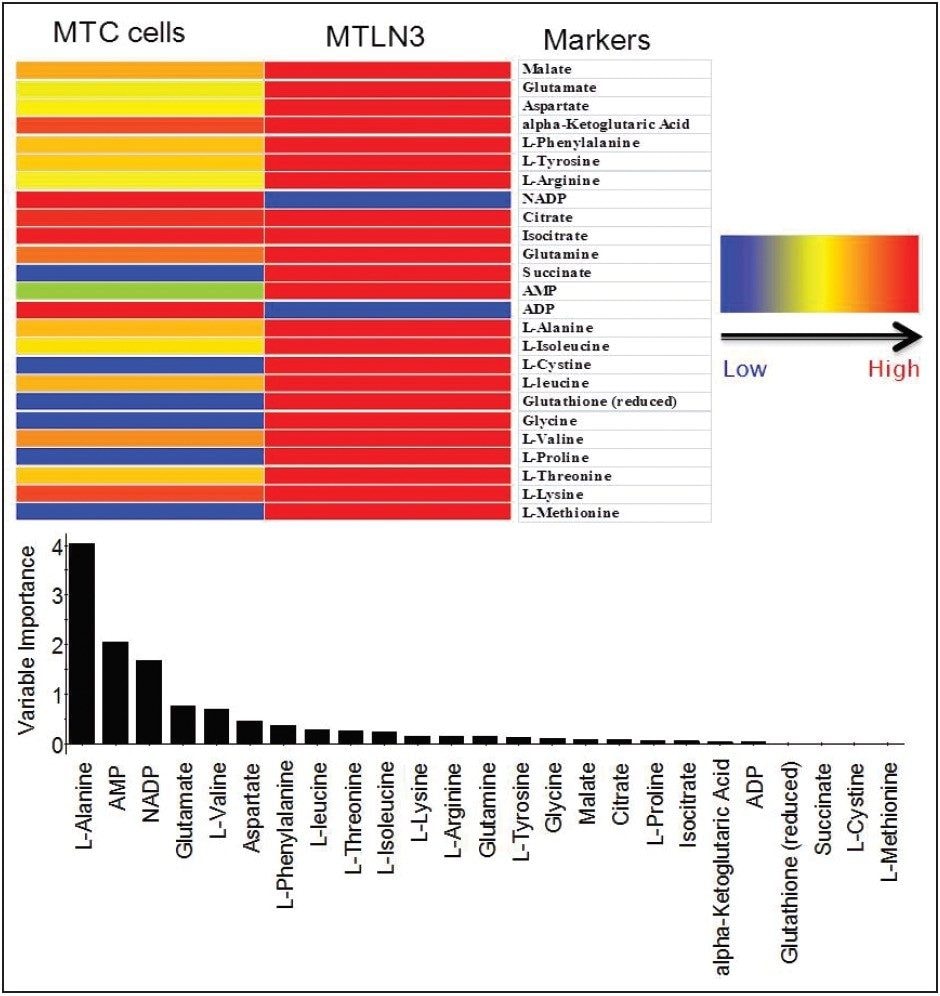
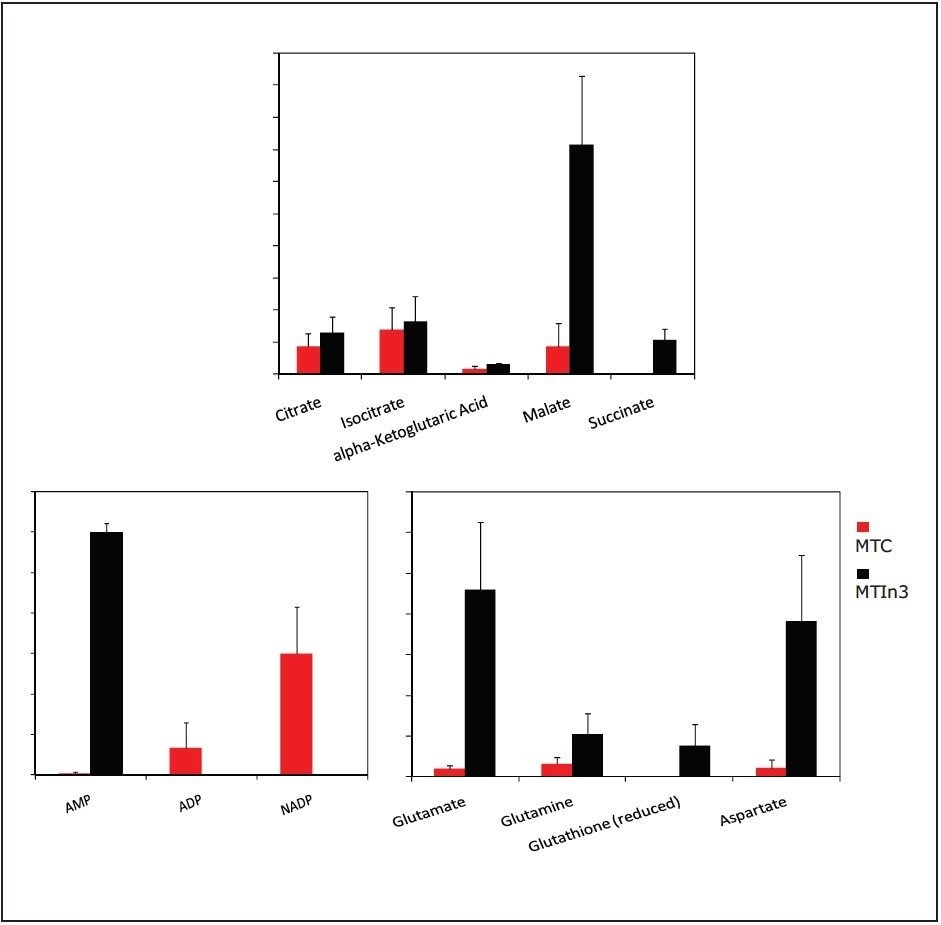
As shown in Figure 6, adenosine monophosphate (AMP) is extremely high in concentration. This indicates that the highly aggressive MTln3 cells require a large amount of energy. Even with a high malate/aspartate shuttle, adenosine triphosphate (ATP) production cannot keep pace.
AMP functions as an energy sensor and regulator of metabolism. When ATP production does not keep up with needs, a higher portion of a cell’s adenine nucleotide pool is made available in the form of AMP. AMP then stimulates metabolic pathways that produce ATP in the MTln3 cells.
The “signature” of high levels of malate, glutamate, aspartate, and alpha ketoglutarate (as high as 10 fold) means high cytosolic NADH must use these carriers for transport into the mitochondria to turn into ATP. Highly aggressive cancer cells, such as MTln3, have high glycolysis and need the malate/aspartate/glutamate/ alpha-ketogutarate shuttle system to satisfy the ATP needs. This shuttle cannot work fast enough because AMP is still very high.
For untargeted global profiling analysis, all the samples were run in triplicate. A QC sample was made by mixing equal volumes of each sample. 10 QC samples were injected prior to the first sample in the experiment. A QC sample was also injected every 10 sample injections.
For the data analysis, MarkerLynx XS Application Manager3 was used to integrate and align chemical and biological MS data points and convert them into Exact Mass Retention Time (EMRT) pairs. Those EMRT pairs can then be used for multivariate statistical analysis, such as principle component analysis (PCA-X), partial least-squares to latent structures data analysis (PLS-DA), and orthogonal PLS data analysis (OPLS-DA) to visualize and interpret the information-rich and complex MS data, as shown in both Figures 8 and 9.
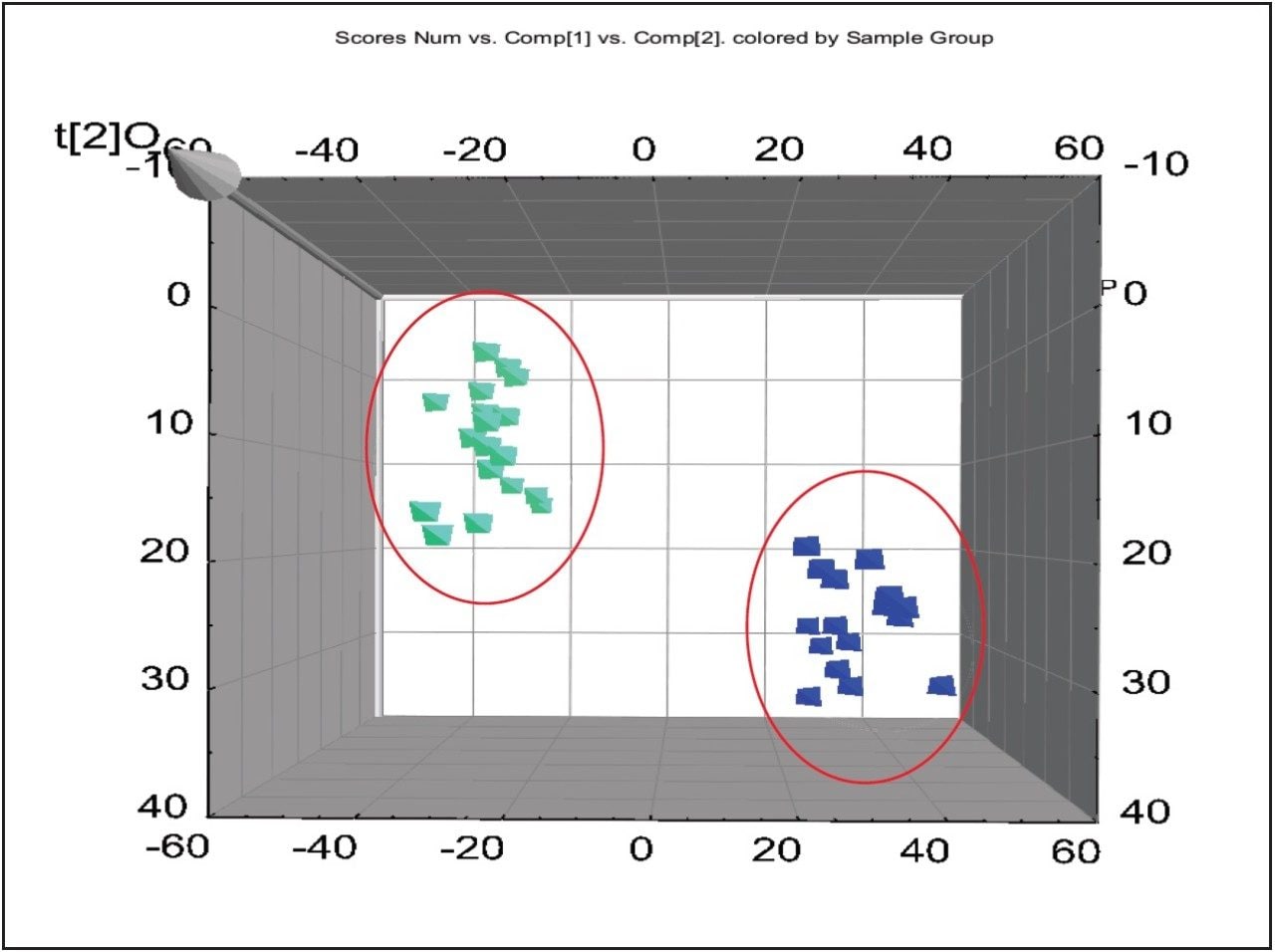
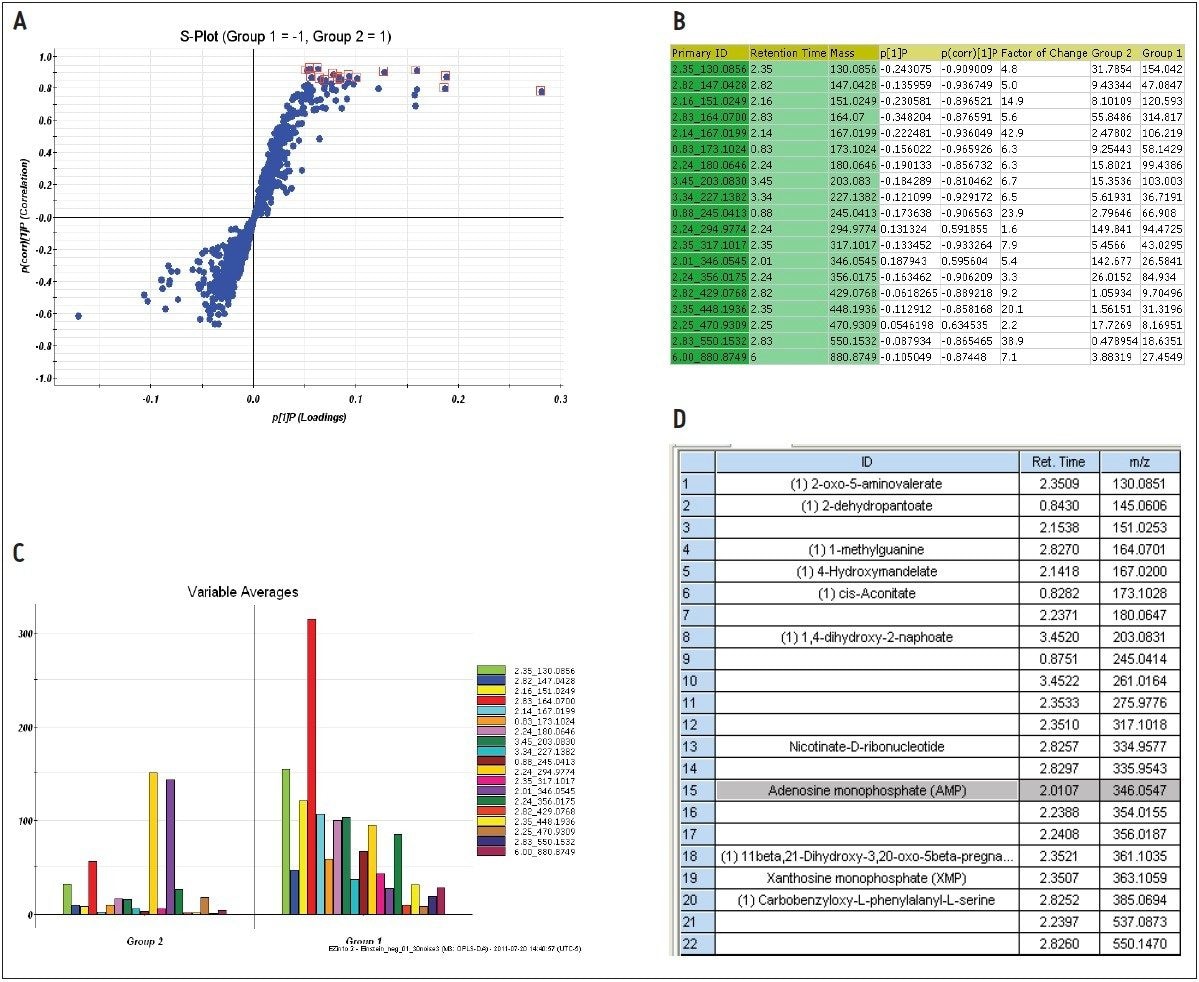
The markers from MarkerLynx statistical analysis were validated, in part, by identifying hits in pathways complementary to those found in targeted analysis, and builds belief in new untargeted hits found. For example, untargeted analysis, shown in Figures 4 and 6, indicated high AMP and phosphoenelpyruvate (PEP) along with high cis-aconitate in aggressive cells. AMP was identified from targeted analysis; cis-aconitate supports targeted analysis finding for increased flux into the TCA cycle, and PEP for increased glycolysis. Markers/carriers (malate/aspartate shuttle) for high cytosolic NADH from targeted analysis are complementary to untargeted findings of high nicotinamide-D-ribonucleotide, a step in NAD synthesis degradation product of amino acids found to be elevated by targeted analysis. Targeted analysis found high levels of aspartate, isoleucine, tyrosine, arginine, and others. Untargeted analysis showed markers for amino acid degradation with high 2-oxo-5-aminovalerate, a breakdown product of arginine; 1,4 dihydrooxy-2-naphoate, a breakdown product of tyrosine; alpha-hydroxyisovalerate, a marker for branched chain amino acid (isoleucine) breakdown; and homoserine, a breakdown product of aspartate.
Among the new markers found from database searching by untargeted analysis are 2-dehydropantoate and 4-hydroxymandelate, as shown in Figure 9D. They are indications of increased methyl transferase activity, which is key to function of biosynthetic pathways. Our results show that any of these pathways appears to be upregulated in the MTln3 cells.
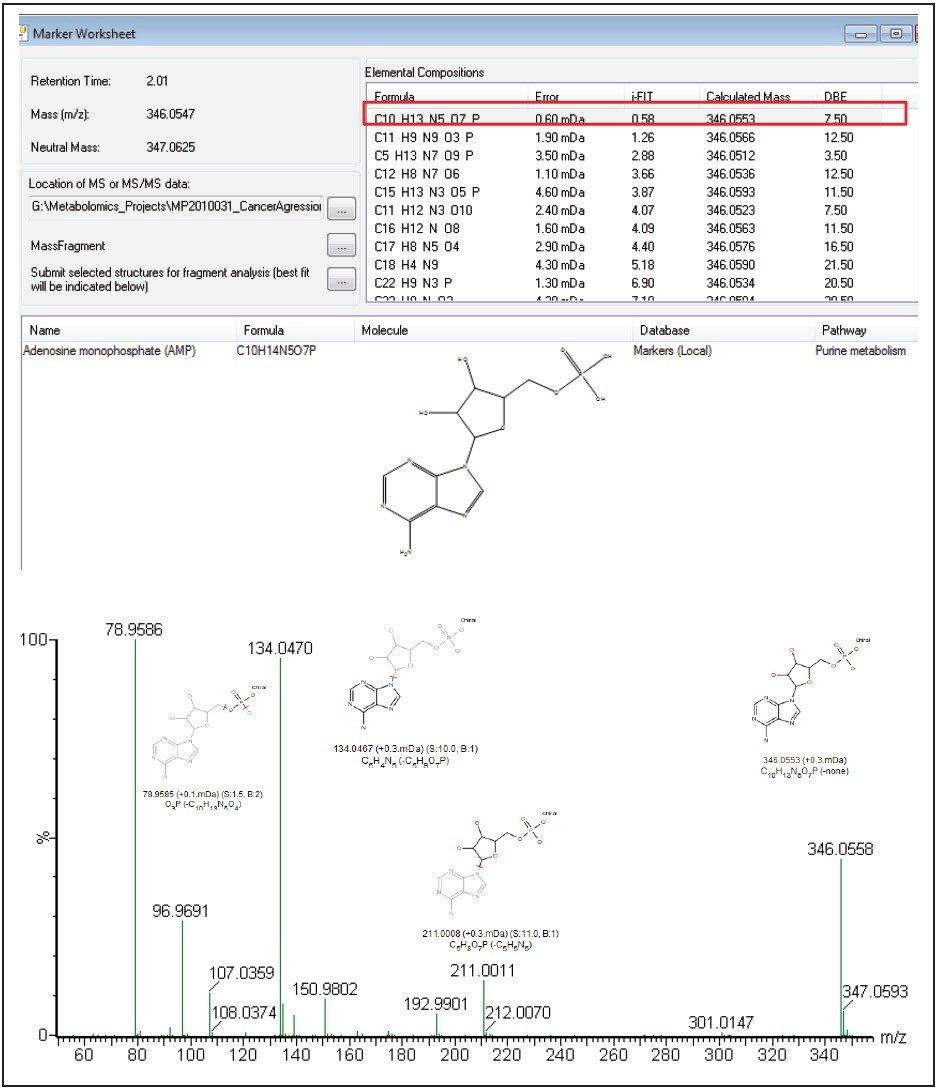
We have successfully demonstrated a metabolomics study workflow that combines targeted and untargeted approaches for breast cancer biomarker analysis.
Aggressive cell MTLn3 and non-aggressive cell MTC show dramatically different concentrations of the biomarkers, such as malate and AMP in glycolysis and TCA cycle, which indicates glycolysis is higher in MTln3 cells.
Known markers of cancer aggressiveness can be analyzed by a targeted approach using Xevo TQ or Xevo TQ-S.
Hits are validated by identifying hits in pathways complementary to those found in targeted analysis using SYNAPT G2; this builds belief in new untargeted hits identified/discovered.
New markers and thereby new pathways can be discovered by untargeted SYNAPT G2 analysis. One example would be 2-dehydropantoate and 4-hydroxymandelate, which are markers for increased methyl transferase activity. Methyl transferase activity is key to the function of biosynthetic pathways.
720004106, October 2011