Qualitative and Quantitative Proteomic Profiling of Cripto-/- Embryonic Stem Cells by Means of LC/MSE Analysis
Introduction
Embryonic stem (ES) cells are attracting significant scientific attention and offer the potential for advanced medical treatment. As such, their characterization and mechanism of differentiation is of great scientific interest. ES cells, deriving from the inner cell mass of the blastocyst, are able to differentiate into all lineage derivatives of the three germ layers: ectoderm, endoderm, and mesoderm, as seen in Figure 1. ES cells are therefore an outstanding model system both for developmental studies as well as for biomedical applications. Recently, particular attention has been paid to molecules and signaling pathways that control the balance between ES cell self-renewal and differentiation. Cripto is a key regulator of ES cell differentiation and mouse Cripto-/- ES cells have been utilized to investigate the molecular mechanisms underlying early events of mammalian lineage specification and differentiation.
A multi-dimensional nanoscale LC/MS/MS approach has been used to qualitatively profile the cripto–/– ES cell proteome. The study enabled a large dataset to be recorded from relatively low sample amounts. Proteins identified by databank searching have been classified in terms of subcellular localization, molecular function, and biological process as defined by their associated gene ontology annotation. A quantitative profile of the cripto–/– ES cells’ proteome was obtained by performing a label-free quantitative LC/MS experiment, utilizing data independent, alternate scanning LC/MSE. The principle of the method is based upon the measurement and subsequent comparison of the chromatographic peak area for each peptide across samples. These peptides are mapped back to their constituent proteins to determine the relative amounts of each protein. This mapping allows an integral view of the alterations induced in stem cell functions by deleting the cripto gene. Several differentially expressed proteins have been identified and quantified in cripto-/- versus wild type (wt) ES cells. Moreover, details are provided on how LC/MS/MS and LC/MSE can aid in studying ES cell differentiation at both the peptide and protein levels.
Experimental
Cell Cultures and Sample Preparation
Wild type (RI) and cripto-/- ES cells were maintained in their undifferentiated state by culture on mitomycin C-treated mouse embryonic fibroblast feeder layers.
Monolayer cultures of cell lines were harvested, after three washes in ice-cold PBS, by incubation with a solution that contained trypsin and EDTA. After centrifugation, the cell pellets were washed three times with PBS and resuspended in 25 mM NH4 HCO3/0.5% RapiGest™ SF for cell lysis and protein extraction. Samples were then sonicated and centrifuged to eliminate cellular debris. The supernatants were collected and protein concentration determined by the Bradford method.
Total protein extracts were subsequently reduced (10 mM DTT), alkylated (10 mM IAA), and enzymatically digested with trypsin, 1:50 (w/w) enzyme:protein ratio.
LC/MS Conditions
The qualitative online 2D-LC/MS/MS setup employed a 180 µm x 23 mm SCX column packed with 5 µm polysulfoethyl A for the first dimension separation. A combined salt and organic step gradient was applied to the SCX column by sequentially injecting a series of solvent plugs (20 mM to 200 mM ammonium formate pH 3.2, with 5% to 20% CH3 CN) onto the SCX column. A Symmetry® C18 5 µm, 180 µm x 20 mm reversed-phase trap column was used both to collect the peptides that elute from the first dimension SCX column for the qualitative study and for preconcentration and sample desalting for the quantitative profiling study.
The second dimension qualitative separations and single dimension quantitative LC/MS experiments were conducted using a 1.5 hr reversed-phase gradient from 55 to 40% acetonitrile (0.1% formic acid) at 250 nL/min on a nano ACQUITY UPLC® System. An Atlantis® C18 3 µm, 75 µm x 15 cm nanoscale LC Column was used. Typical on-column sample loads were 2.0 and 0.5 µg of protein digest for the qualitative and quantitative studies, respectively.
Data directed analysis and data independent, alternate scanning LC/MSE experiments were performed with a SYNAPT™ MS Mass Spectrometer.
2D Gel Electrophoresis
Wild type (RI) and Cr–/– ES cells protein extracts were separated in the first dimension on a non-linear pH 3 to 10 gradient, and in the second dimension on homogeneous polyacrylamide gels. The gels were stained with colloidal Coomassie blue stain, followed by protein identification using a MALDI LR™ Mass Spectrometer.
Results and Discussion
Qualitative Profiling
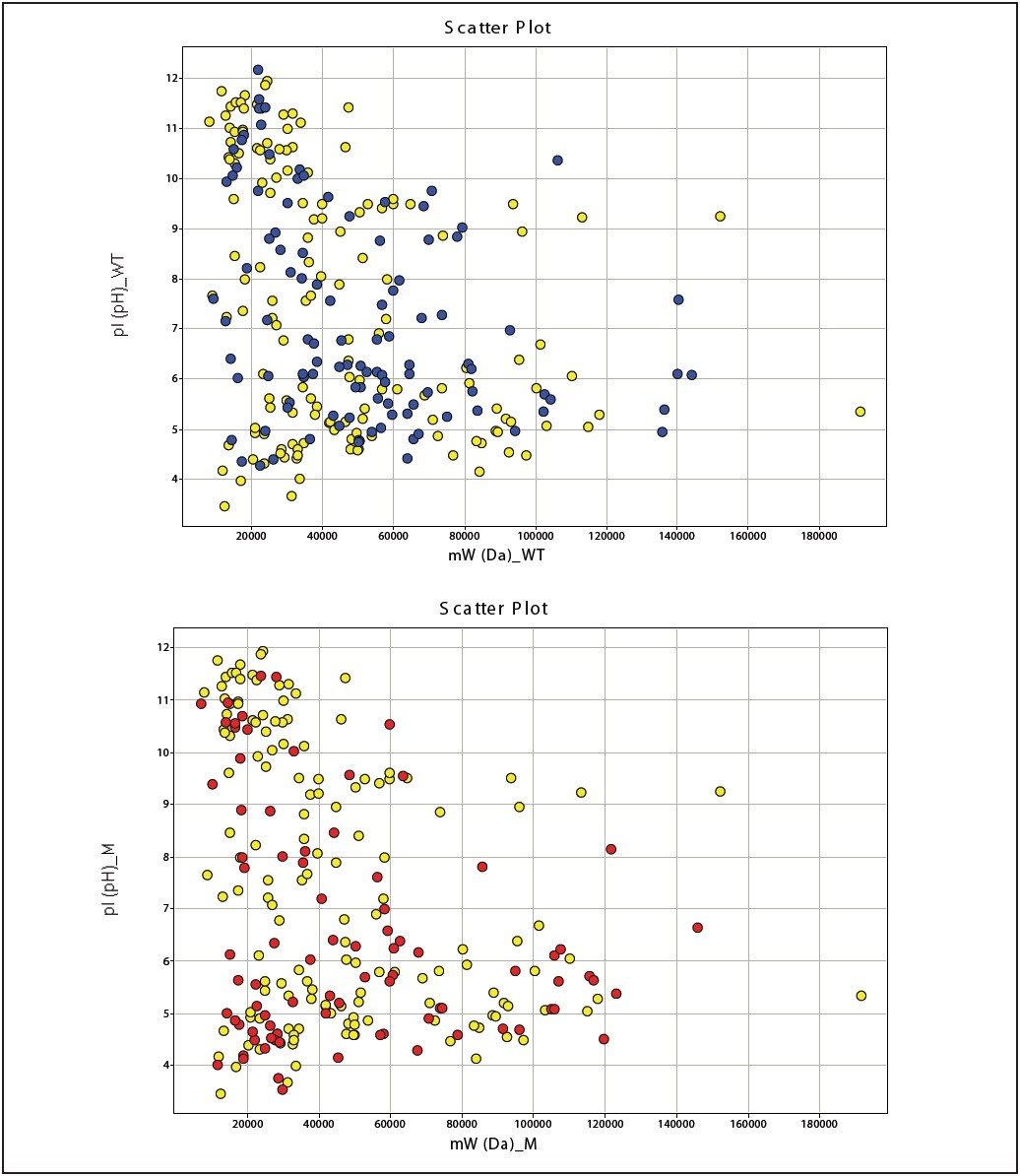
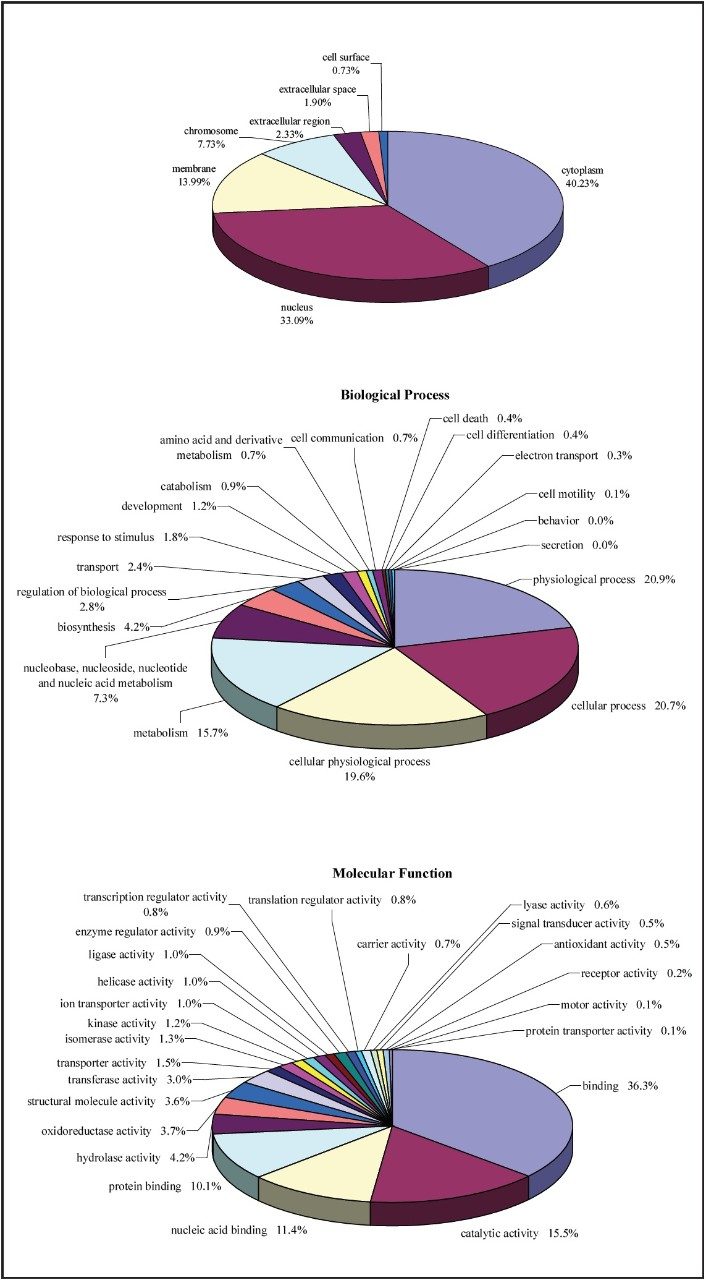
The database search results of the 2D-LC/MS/MS data for both cell lines were used to generate qualitative profiles, as shown in Figure 2, and to conduct Gene Onthology (GO) annotation experiments, as shown in Figure 3. 106 and 85 proteins were identified to the IR and Cr-/- cell lines, respectively, and 146 proteins were identified commonly to both cell lines.
Quantitative Profiling
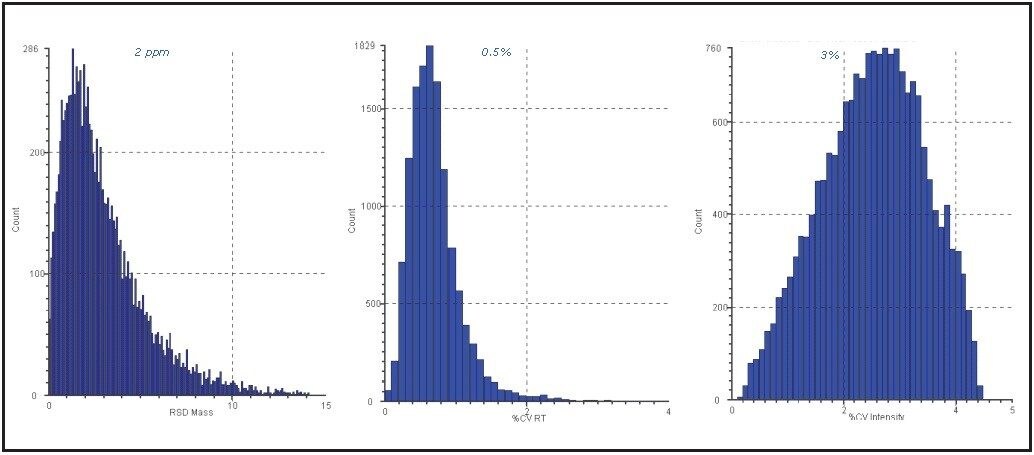
The analytical reproducibility was assessed prior to conducting label-free quantitative analysis. The figures of merit for one of the investigated conditions are shown in Figure 4.
Quantitative analyses were conducted at both the protein and peptide levels. Figure 5 demonstrates an example of filtering on the basis on the probability of peptide regulation.
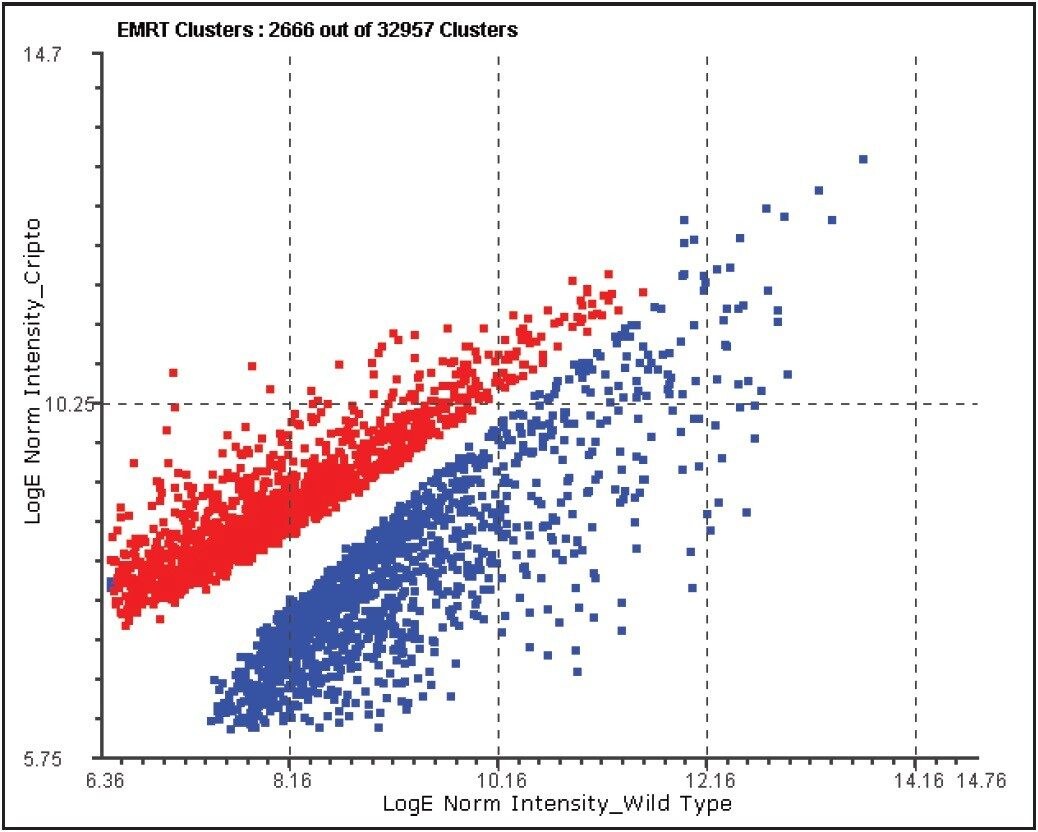
Further filtering of the quantified proteins was conducted on the basis of replication (≥2 out of 3 replicate injections), significance of regulation (probability of regulation >95%), and regulation coefficient of variation (CV<0.02). An example of exclusively upregulated proteins in the cripto-/- cell line is shown in Table 1.
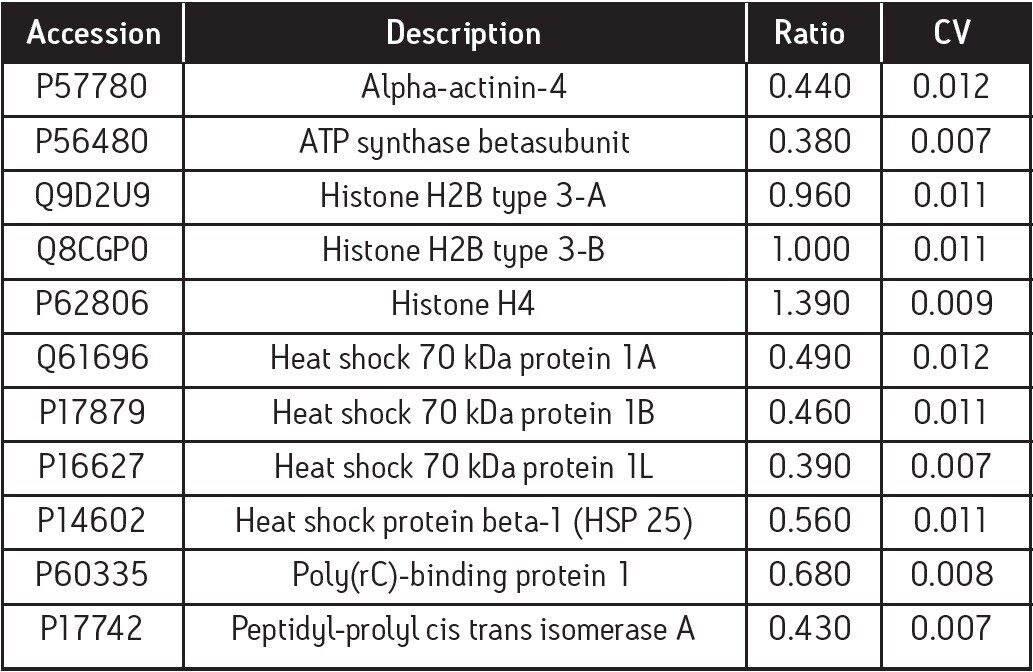
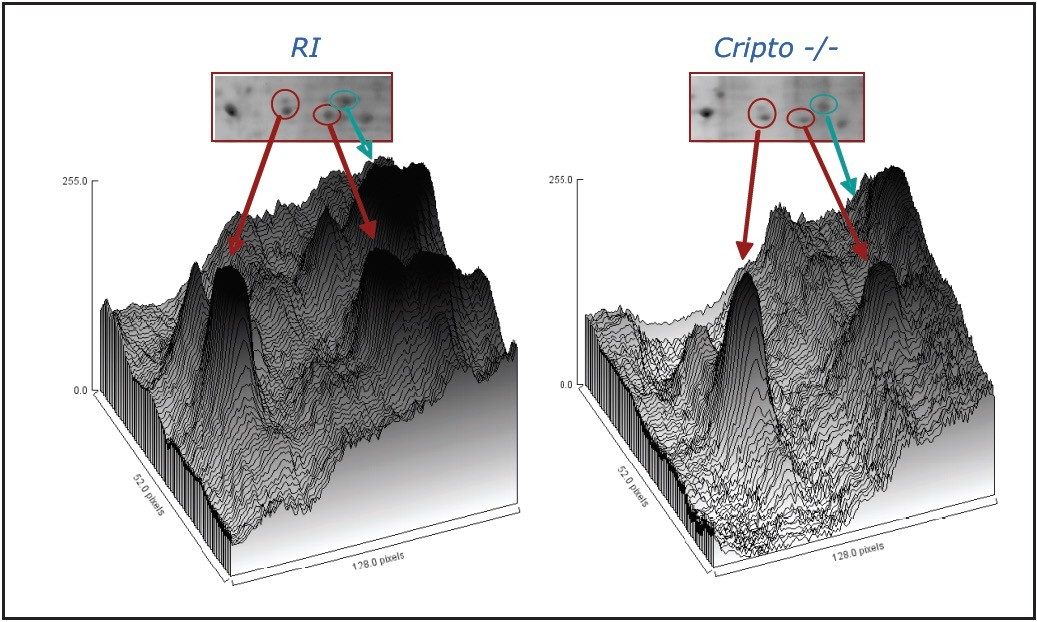
As an example, HSP25 is essential for functional differentiation into beating cardiomyocytes, but is not required for neuronal differentiation and has been positively identified by quantitative label-free LC/MS and 2D gel analysis, as shown in Table 1 and Figure 6.
Conclusion
- Proteomic profiling of cripto-/- and wild type embryonic mouse stem cells by means of LC/MS, utilizing 2D nanoscale LC/MS/MS and data-independent, alternate scanning LC/MSE, has been shown
- Label-free LC/MSE analysis has been performed to obtain a quantitative comparison of proteins differentially expressed, and to complement the qualitative 2D nanoscale LC/MS/MS view of the proteomes of the two cell lines
- Both qualitative and quantitative information was obtained within a single LC/MSE experiment, which cannot be as readily achieved with labeled-oriented quantitative methods, since additional sample preparation would be required
- In total, around 300 proteins have been identified in each separate sample and subsequently quantified. In comparison to more traditional biochemical protein quantification techniques, such as ELISA, Western blotting, etc., this provides a more parallel and unbiased approach towards protein quantification
- Large fractions of the protein population were expressed univocally in the wild type or the cripto-/- cell lines
- Quantitative MS results were in agreement with proteins described in literature that are affected by cripto ablation and 2D gel analysis
- The obtained data indicates that a hallmark of neuronal differentiation was already established in undifferentiated ES cells; hence, in the absence of cripto
References
- Bradford MM. A Rapid and Sensitive Method For the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal Biochem. 72: 248–254. 1976.
- Silva JC, Denny R, Dorschel CA, Gorenstein M, Kass IJ, Li GZ, McKenna T, Nold MJ, Richardson K, Young P, Geromanos S. Quantitative Proteomic Analysis by Accurate Mass Retention Time Pairs Anal Chem. 77 (7): 2187–2200. 2005.
- Silva JC, Gorenstein MV, Li GZ, Vissers JPC, Geromanos SJ. Absolute Quantification of Proteins by LCMSE; A Virtue of Parallel MS Acquisition Mol. Cell. Proteomics. 5(1): 144–156. 2006.
- Battersby A, Jones RD, Lilley KS, McFarlane RJ, Braig HR, Allen ND, Wakeman JA. Comparative Proteomic Analysis Reveals Differential Expression of HSP25 Following the Directed Differentiation of Mouse Embryonic Stem Cells. Biochim Biophys Acta. 1773(2): 147–156. 2007
- Winger QA, et al, Heat Shock Protein 1 and the Mitogen-Activated Protein Kinase 14 Pathway Are Important for Mouse Trophoblast Stem Cell Differentiation. Biol. Reprod. 76(5): 884–891. 2007.
- Parisi S, D’Andrea D, Lago CT, Adamson ED, Persico MG, Minchiotti G. Nodaldependent Cripto Signaling Promotes Cardiomyogenesis and Redirects the Neural Fate of Embryonic Stem Cells. J. Cell Biol. 163 (2): 303–314. 2003.
- Minchiotti G. Nodal-dependant Cripto Signaling In ES Cells: From Stem Cells to Tumor Biology. Oncogene. 24 (37): 5668–5675. 2005.
Featured Products
720002990, March 2009