
This application outlines a UPLC method for the separation/purification of double-stranded DNA (dsDNA) sequences ranging from 50 to 600 base pairs in length, in under 20 minutes.
Over the past 20 years there has been a considerable amount of effort focused on the determination of detailed maps of genomes from various species and individual genome analysis. This work is leading to an increased understanding of susceptibility to disease and provides putative sequence targets for oligonucleotide-based therapeutic strategies.
In general, molecular biology methods for manipulation of DNA rely on restriction enzymes, polymerase chain reaction (PCR), and sequencing techniques. Using these methods, genomic DNA is typically converted into shorter double-stranded (ds) DNA sequences, typically 100 to 1000 base pairs (bp) in length. The shorter dsDNA molecules are often analyzed or isolated by methods such as slab gel or capillary electrophoresis and anion-exchange LC. The method outlined in this application note uses a volatile ion-paring system, reducing post-purification processing.
Besides gel electrophoresis, Waters UltraPerformance Liquid Chromatography (UPLC) Technology can also be used for rapid and cost-effective separation and purification of wide array of dsDNAs.
This application outlines a UPLC method for the separation/ purification of dsDNA sequences ranging from 50 to 600 base pairs in length, in under 20 minutes. The separation is based on DNA length, rather than on the sequence. Because of the substantial column mass load capacity, large amounts of dsDNA fragments can be isolated.
This presented method dramatically reduces analytical time and effort compared to gel electrophoresis, and can be utilized for separation of dsDNA fragments produced by hydrodynamic shearing. Additionally, isolated samples may be used for next-generation sequencing. With current research efforts focused on reducing total genome processing times and sequencing costs, the Waters ACQUITY UPLC System solution provides for a superior tool to those currently used for dsDNA isolation, purification and analysis.
The HaeIII digest of pBR322 plasmid was purchased from Sigma-Aldrich. MspI digest of pBR322 was obtained from New England Bio Labs. These digests were chosen because they contain few dsDNA fragments of the similar or same length, but cover a broad range of dsDNA lengths. The stock solutions were diluted 1:10 in 100 mM triethylammonium acetate (TEAA). Typical injection volumes were 10 μL, giving on column loads of 0.6 μg.
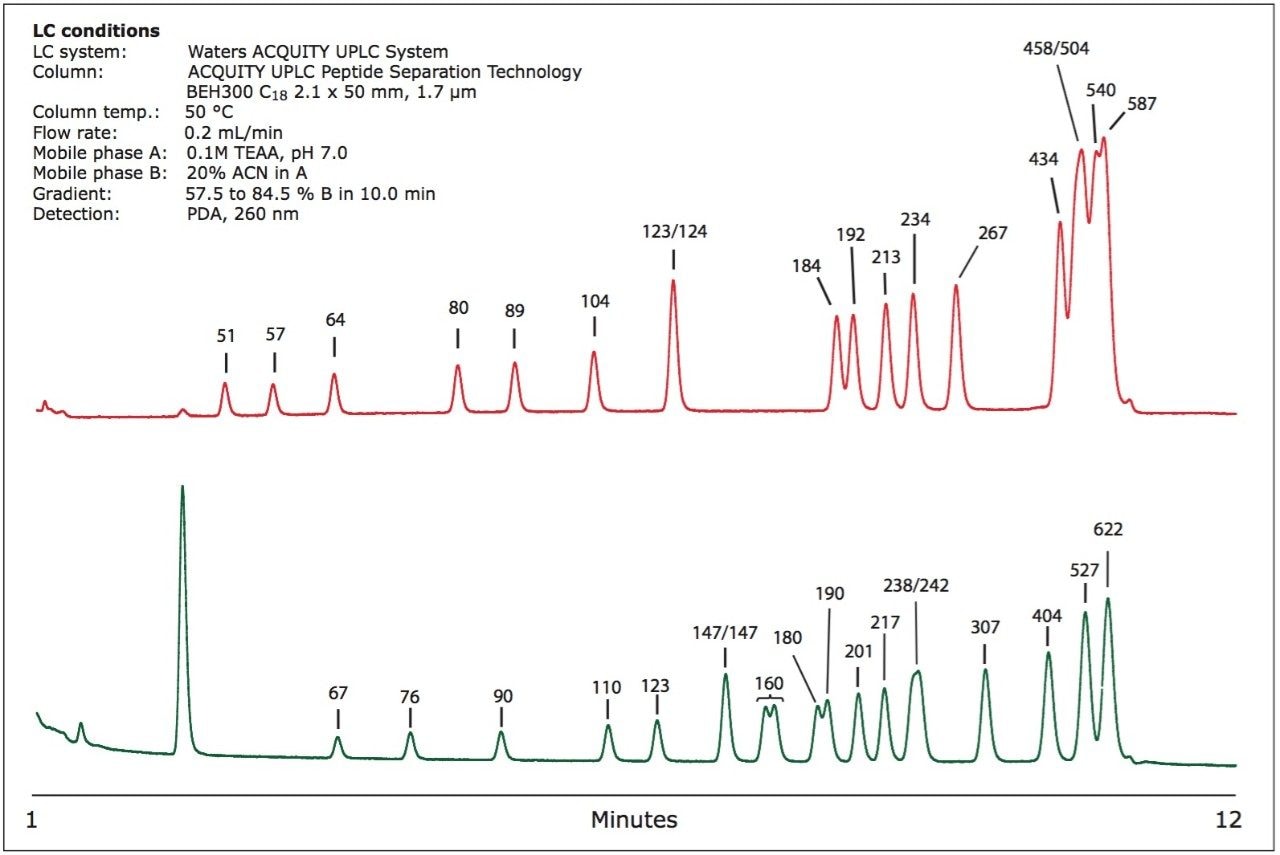
dsDNA mixtures were separated using the ACQUITY UPLC System with an ACQUITY UPLC Peptide Separation Technology BEH300 C18 2.1 x 50 mm, 1.7 μm column using ion-pair, reversed-phase chromatography.1 The pore size of the sorbent was 300 Å. Separated products were detected with a Waters ACQUITY UPLC PDA detector at 260 nm. Mobile phase A consisted of 0.1 M triethylammonium acetate (TEAA); mobile phase B was composed of 20 % acetonitrile in mobile phase A. The column temperature was maintained at 50 °C. Gradient was 57.5 to 84.5 % B (11.5 to 16.9% ACN) in 20 minutes.
The BEH300 column material was selected for the dsDNA separation for several reasons. First, BEH Technology offers exceptional stability, allowing for use of a single column over the long term. Secondly, the 300 Å pore size allows for an efficient separation of longer dsDNA fragments. This is primarily due to the increased accessibility of pores to large molecular weight analytes.
As shown in Figure 1, the UPLC separation provides good resolution of dsDNA, especially in the 50 to 300 base-pairs (bp) region. Eluting peaks can be easily collected, unlike using gel electrophoresis where the band needs to be excised and isolated from the gel and desalted.

To determine whether the UPLC separation strategy provided predictable dsDNA retention, we plotted retention time vs. oligonucleotide length for the HaeIII digest. The data were fit to an inverse third order polynomial, Equation 1, yielding an excellent correlation coefficient of 0.9999.
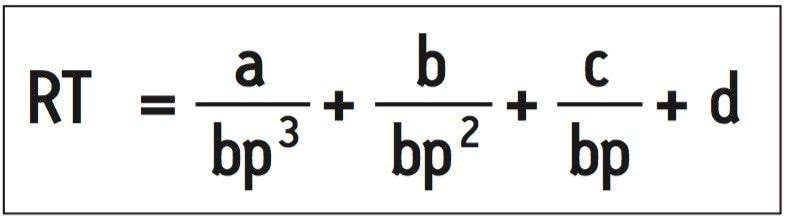
Additionally, we plotted and fit the MspI data in the same manner yielding a correlation coefficient of 0.9996 (curve not shown). Excellent correlation between the expected and observed retention times was found regardless of which digest was used for curve fitting.
This correlation strongly indicates that our separation system provides exemplary resolution of dsDNA sequences, up to 600 bp, and provides a predictable elution order regardless of oligonucleotide sequence.
Accuracy in retention time prediction allows for the collection of fractions of specific lengths if desired. In order to investigate the sizing accuracy of the UPLC method, we calculated the expected bp length from the retention times for the MspI digest using the curve for the HaeIII digest (Figure 2). These values were compared to the actual bp lengths and expressed as absolute and percent bp error.

As shown in Figure 3, there is very good correlation between the expected and observed bp lengths. The red line indicates that there is less than 5% variation in the predicted oligonucleotide length compared to the fitted curve (fitted for HaeIII data). This error translates to less than a 10 bp error for oligonucleotide lengths up to 400 bp.
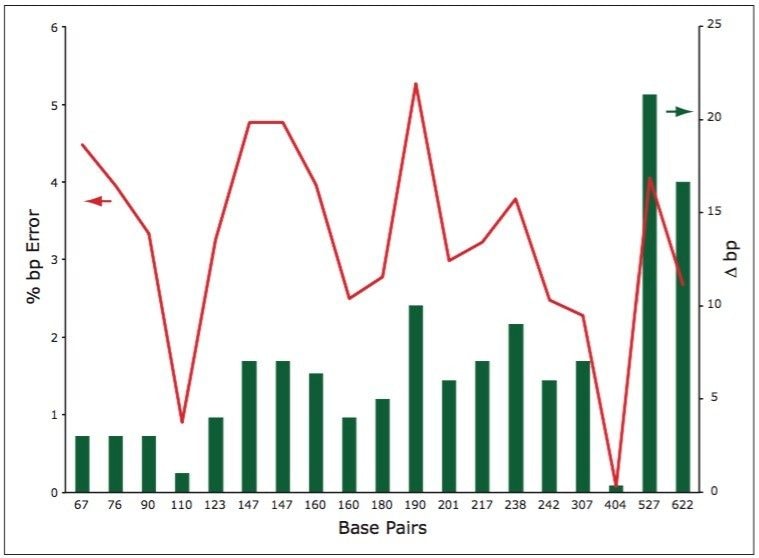
One can note that there is more significant absolute deviation for 527 and 622 bp fragments (15 to 22 bp), but the relative difference remains below 5%. This sizing accuracy is acceptable for many molecular biology applications.
Some deviation in retention behavior was detected for shorter dsDNA, generally below 40 bp in length (Figure 2). This is due to gradient delay effect at the beginning of analysis. It is advisable to start the gradient with lower elution strength solvent when working with shorter dsDNA fragments.
Additionally, there appears to be a limited contribution of oligonucleotide dsDNA sequence to the observed retention time. Although this effect is limited, it can be observed as imperfect co-elution of dsDNA fragment of the same length, but different sequence. While minor sequence contribution was observed, it is clear from our data that elution in UPLC correlates very closely to dsDNA length.
We also investigated the utility of our method using more rapid chromatographic separations. To accomplish this we increased the gradient slope by a factor of two at constant flow rate. As shown in Figure 4, there is a slight loss in resolution, primarily for oligonucleotide lengths above 300 bp, however very good resolution for moderate length oligonucleotides is achieved
The dsDNA separation strategy utilizing the ACQUITY UPLC System presented here provides an accurate, high throughput, and reproducible method that allows for the prediction of retention time for a desired oligonucleotide length, and collection of oligonucleotide lengths of interest.
The method can be scaled by using larger column configurations available from Waters, which is useful for the researcher seeking to do large-scale separations of dsDNA.
When combined, the accuracy, high throughput, and ease of this method offer a significant advantage other methods currently in use.
As illustrated, this UPLC method allows separations to be performed at a variety of gradient slopes, allowing for analysis and sample collection at a variety of timescales, depending on the needs of the researcher, with minimal loss of resolution for moderate length oligonucleotides.
This powerful separation solution relies on the outstanding stability and reproducibility offered by Waters BEH column chemistry. Following peak collection, samples can be aliquoted and dried for long-term storage. The volatility of TEAA allows for an easy removal of ion-pairing buffer components, yielding oligonucleotides that are practically salt-free and suitable for storage as necessary, offering another advantage over currently used methods.
720002741, July 2008