Digesta Ex Machina: Automating Sample Preparation for Oligo Mapping With Rapizyme™ MC1 and Cusativin on Andrew+™ Robot
Abstract
This study was designed to demonstrate the suitability of the Andrew+ Pipetting Robot for automation of the RapiZyme MC1 and Cusativin digestion of RNA. The manual protocol was adapted for reproducible RNA digestion on the automated platform with minimal method deviations. Here, we showcase the equivalency of manual and automated preparations for digestion of sgRNA and the reproducibility of the Andrew+ Pipetting Robot protocol.
Benefits
- Simple, straightforward procedure is readily automatable for implementation in quality control (QC), analytical development, and research environments
- Scalable automated sample preparation for digestion of eight RNA samples in under 45 minutes
- Reproducible digestions of RNA ready for direct LC-MS analysis without the need for inhibitors or additional clean up steps
- Achieve higher sequence coverage with RapiZyme MC1 and Cusativin providing longer overlapping, LC-MS amenable digestion products from partial digestion
Introduction
Background
RNA therapeutics have great potential to transform medicine and combat a range of incurable diseases. RNA can be produced synthetically (CRISPR sgRNA) or through in vitro transcription (mRNA), requiring advanced characterization of CQAs: purity, structural integrity, identity, sequence modifications, safety, and efficacy. Due to its size, long RNA require digestion with endonucleases before analytical characterization by LC-MS. The resulting digestion products can be effectively analyzed using ion-pairing reversed-phase (IPRP) or hydrophilic interaction liquid chromatography (HILIC) coupled with mass spectrometry (MS).
Waters RapiZyme MC1 and RapiZyme Cusativin provide controlled and reproducible RNA cleavage with targeted dinucleotide specificities.1 The unique cleavage patterns produced by RapiZyme RNases generate a variety of RNA digestion products with overlapping sequences that facilitate sequence analysis of RNA and results in greater sequence coverage. While this application note highlights the automation of RapiZyme RNases for digestion of sgRNA, other RNAs such as messenger RNA (mRNA) or other single stranded RNA may be applicable as well with an optimized protocol.
Prior to analysis, the RNA samples, digestion buffer, and reconstituted enzyme are combined and incubated at 30 °C to perform RNA digestion. After digestion is completed, the enzyme is heat inactivated and the sample was transferred from a PCR tube to a QuanRecovery™ with MaxPeak™ HPS 12 x 32 mm Screw Neck Vial, 300 µL vial (p/n: 186009186). The procedure for RNA digestion is depicted in Figure 1. Here we demonstrated the use of an automated platform Andrew+ Robot with minimal method deviations from manual protocol to reproducibly perform RNA digestions for upstream analysis of RNA therapeutics.
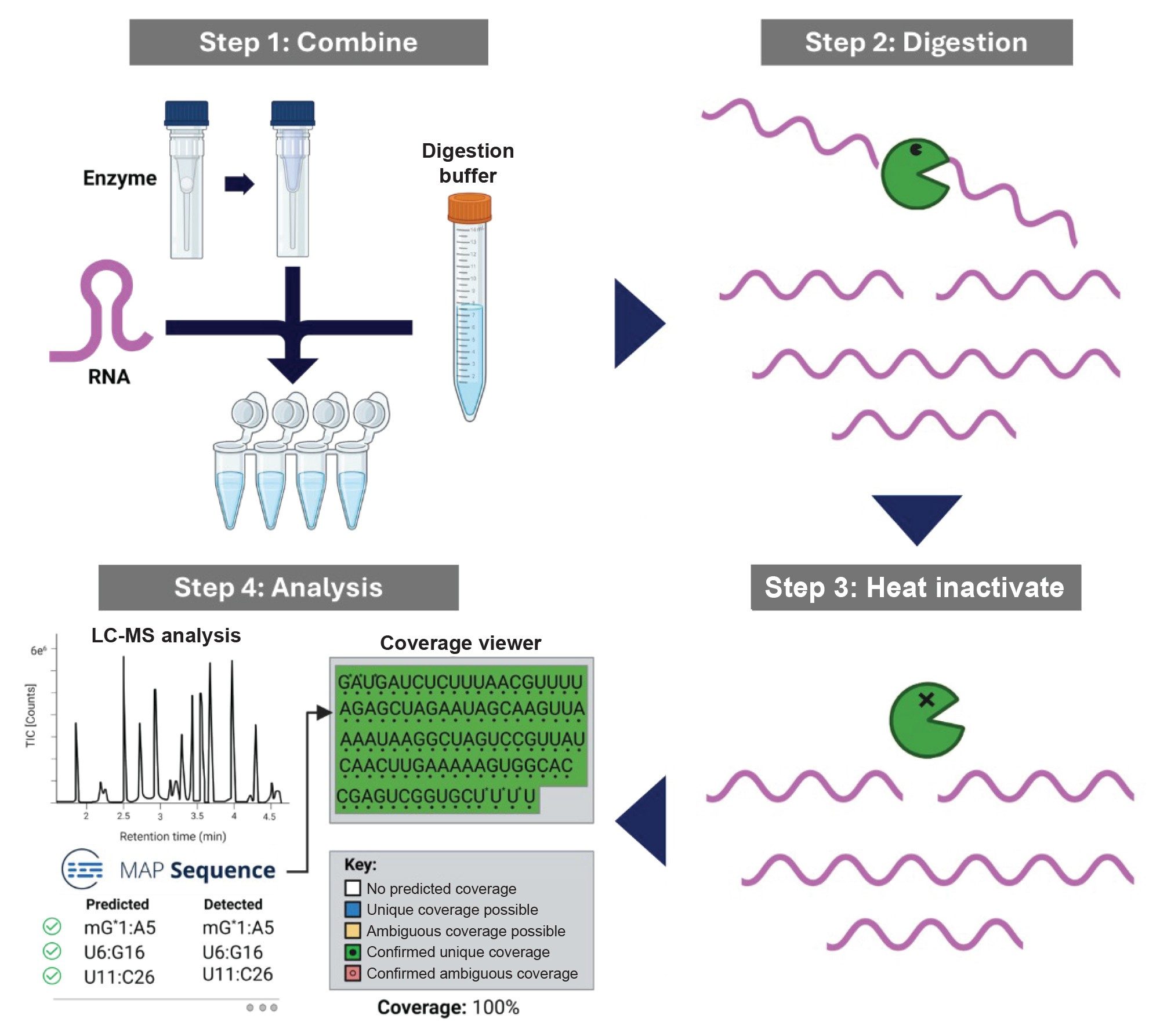
Experimental
Experimental Table
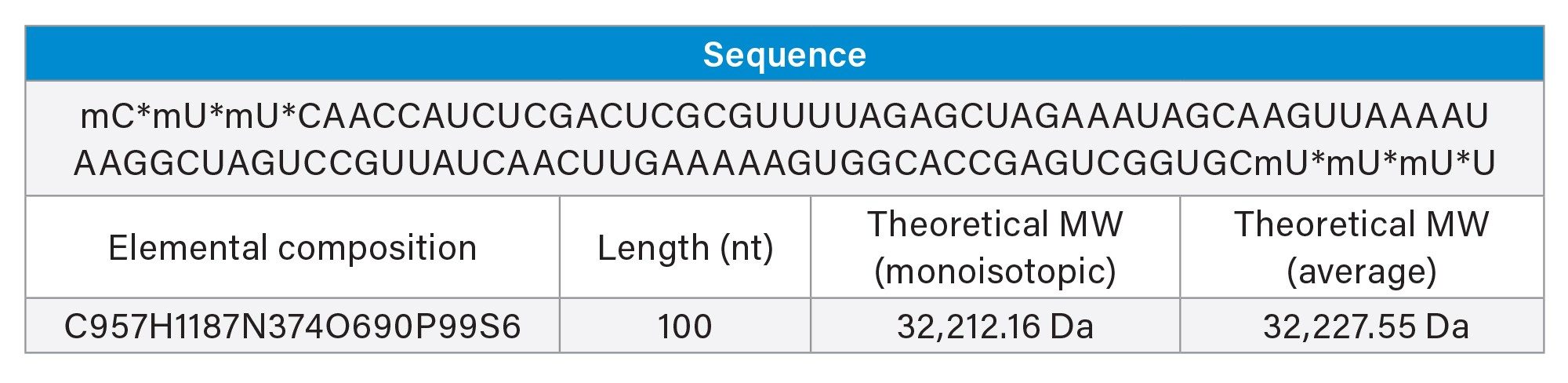
When working with RNA, it is important to follow sterile laboratory techniques to prevent nonspecific cleavages and degradation. All reagents used were RNase-free certified.
Digestion Buffer Preparation
A 25 mL of 200 mM ammonium acetate buffer stock solution was prepared by diluting 1 mL of 5 M ammonium acetate (Invitrogen, AM9071), with 20 mL of RNase free water (Invitrogen, UltraPure distilled water, 10977–015). The pH was adjusted dropwise with ammonium hydroxide (Fisher Chemical, Trace Metal grade, A512–P500), to the appropriate pH (8.0 for MC1 and 9.0 for Cusativin). The final volume was brought to 25 mL with RNase free water and 10 mL of each respective to enzyme stock solution was transferred to a 15 mL centrifuge tube for the automation protocol. The remaining stock was utilized for reconstituting the lyophilized enzyme and saved for future methods.
Enzyme Preparation
One vial of each RapiZyme RNase MC1 (p/n: 186011190) and Cusativin (p/n: 186011192) was reconstituted with 200 µL of 200 mM ammonium acetate buffer at pH 8.0 and pH 9.0, respectively, yielding enzyme stock solution of 50 U/µL. The enzymes were stored in their original skirted tube for the automation protocol.
sgRNA Sample Preparation
The sample utilized for this procedure was 1 nanomole of sgRNA with a sequence that targets the mouse GATA2 gene, a zinc-finger transcription factor. The vial content (32 µg, sufficient for 3 reactions of 10 µg of RNA per single reaction) was reconstituted in 48 µL of RNase free water, gently vortexed, and centrifuged to ensure mixing. Multiple vials were prepared and RNA pulled together for the manual and automated procedures. 15 µL of sgRNA (10 µg) was pipetted into each PCR tube (FrameStrip 8-well PCR tube strip with caps (Azenta, 4ti-0786/R) in 96x FrameStrip adapter (Azenta, 4ti-0370) prior to digestion.
Ion Pairing Reversed Phase Chromatography
|
Column: |
ACQUITY™ Premier Oligonucleotide BEH C18, 300 Å, 1.7 µm, 2.1 x 50 mm (p/n: 186010539) |
|
Mobile phase A: |
0.1% N,N-diisopropylethylamine (DIPEA) and 1% (v/v) IonHanceHFIP (p/n: 186010781) in 18.2 MΩ water |
|
Mobile phase B: |
0.0375% DIPEA and 0.075% (v/v) IonHance HFIP (p/n: 186010781) in 55:10:35 Acetonitrile:Methanol:18.2 MΩ water |
|
LC system: |
ACQUITY Premier System with BSM |
|
Detector: |
Xevo™ G3 QTof Mass Spectrometer |
|
Wavelength: |
254 nm |
|
Flow rate: |
0.4 mL/min |
|
Injection: |
5 µL |
|
Column temperature: |
70 °C |
|
Sample temperature: |
6 °C |
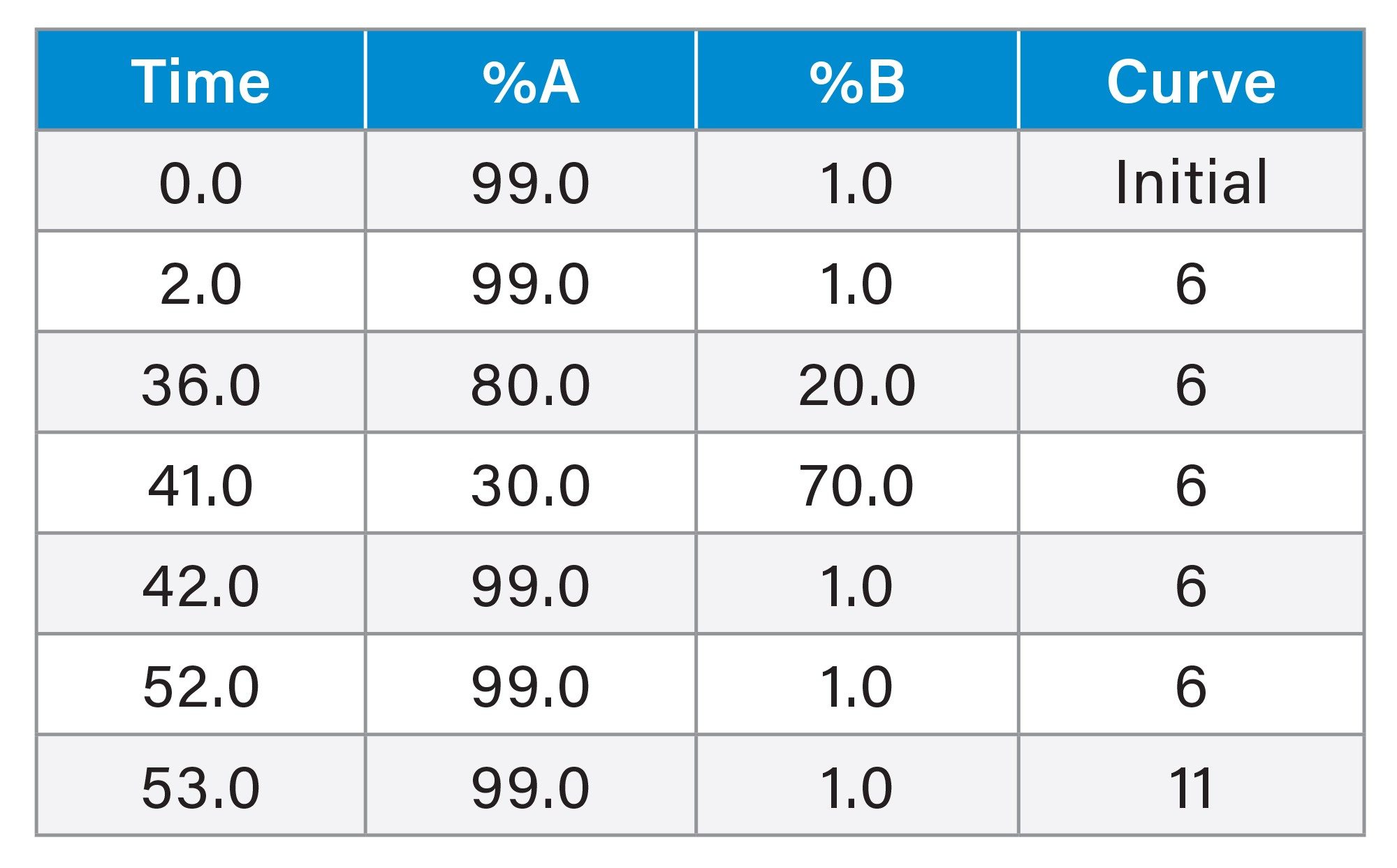
Gradient Table
MS Conditions
|
Source type: |
ESI |
|
Polarity: |
Negative |
|
Analyzer mode: |
Sensitivity |
|
Capillary voltage: |
1.5 kV |
|
Sample cone voltage: |
40 V |
|
Source temperature: |
100 ºC |
|
Desolvation temperature: |
550 ºC |
|
Cone gas: |
50 L/h |
|
Desolvation gas: |
650 L/h |
|
Scan mass range: |
550–2000 |
|
Scan time: |
1.0 sec |
|
Collision energy: Low |
6 V |
|
High energy ramp: |
10–45 V |
|
Intelligent dat capture-intensity threshold: |
Medium (10) |
|
Lock correction: |
Automatic (30 sec intervals) |
|
Events: LC, sample |
Initial-40.0 min |
|
Lock mass: Leucine Enkephalin |
554.26202 (M-H+)1- |
|
Lock mass: combine width and mass window |
3 scans, 0.5 m/z |
Results and Discussion
Labware Considerations for the Manual and Automated Procedure
The same RNase-free PCR tube strips (Azenta, 4-ti-0786) were used for both the manual and automated procedures for comparison. There are other PCR tube and plate formats available in OneLab™ Software, many of which are suitable for this procedure, however their performance was not tested with this automated procedure. The 8-well PCR tube strip with a plate adapter was chosen for its flexibility, as the procedure was specifically designed to analyze eight samples at a time. This format allows for a flexible number of samples to be processed in a single column on PCR plate at a time without having to use an entire 96 well PCR plate for each preparation. For higher-throughput workflows, additional 8-well PCR tube strips or a 96-well PCR plate can easily be adapted as needed. The tubes were purchased with caps which reduce evaporation and the potential for contamination during the incubation steps.
Digestion with MC1 and Cusativin was performed separately as its own standalone protocol. 15 µL of sgRNA stock (0.67 µg/µL) was pipetted into the PCR tubes for the manual and automated preparation. 2 µL of RapiZyme RNase (100 Units) and 13 µL of 200 mM ammonium acetate buffer (pH 8.0 for MC1 and 9.0 for Cusativin) were added to each tube. Incubation was performed for 15 minutes at 30 °C for enzymatic cleavage of RNA and then the reaction was heat inactivated at 70 °C (MC1) and 75 °C (Cusativin) for 15 minutes. The PCR tubes were capped for both procedures prior to incubation to reduce evaporation. The samples were transferred to a microcentrifuge (Eppendorf 5424) and briefly spun down at 10,000 RCF for 10 seconds to recover condensate formed on the tube wall or caps. Following centrifugation, the tubes were uncapped and 25 µL of digestion mixture was transferred to 300 µL QuanRecovery Screw Neck Vials (p/n: 186009186) for LC-MS analysis.
During the manual procedure, the samples were pipetted with Sartorius Taccta Mechanical pipettes into the PCR tube strips and incubated in an Eppendorf Thermomixer C. The automated protocol utilizes Waters Electronic Pipettes, and a high-speed Peltier+ connected device formatted for PCR tubes for incubation steps. Both workflows include the same centrifugation step. All other labware, reagents, and pipetting volumes were kept the same for performance comparison.

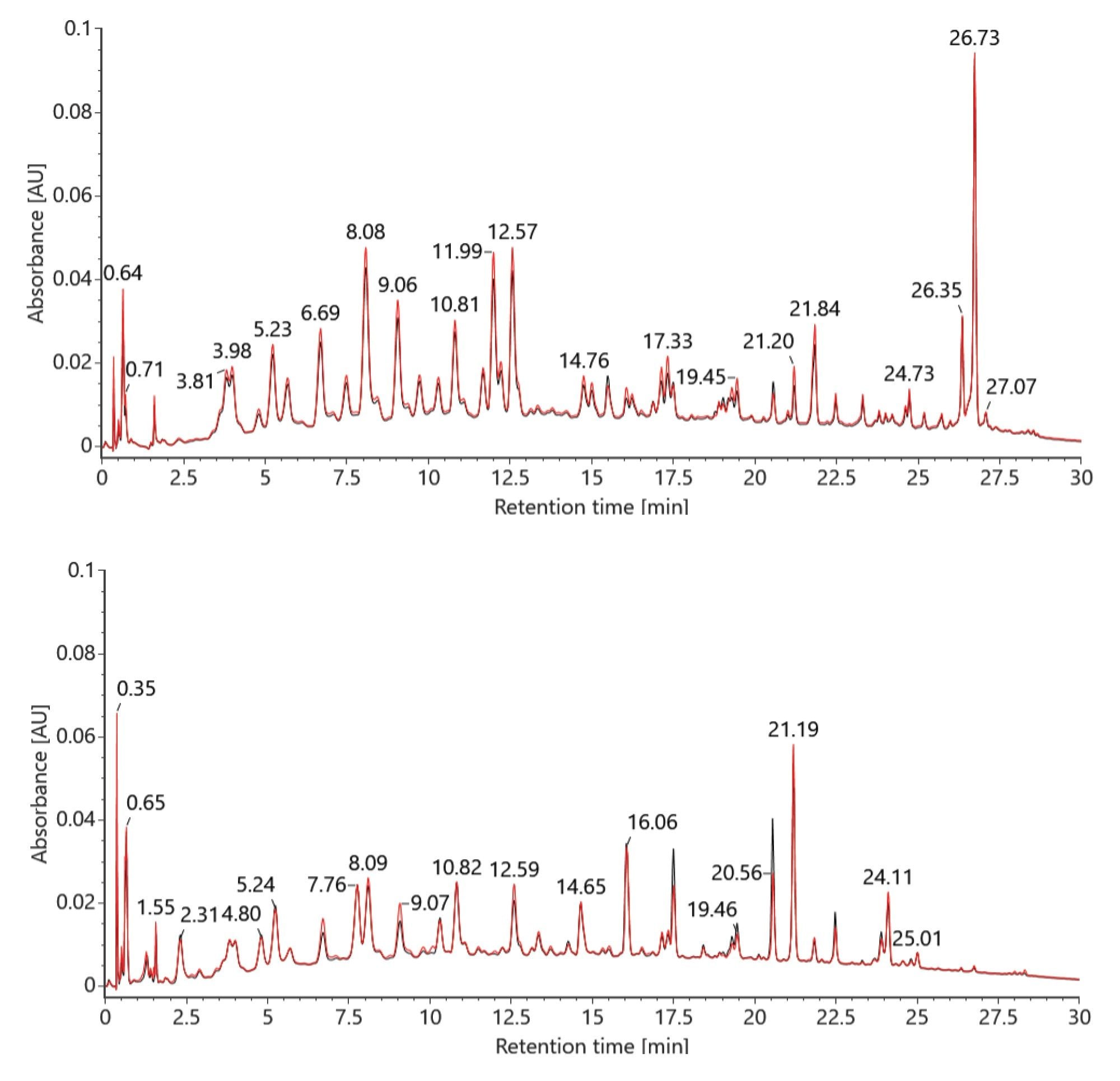
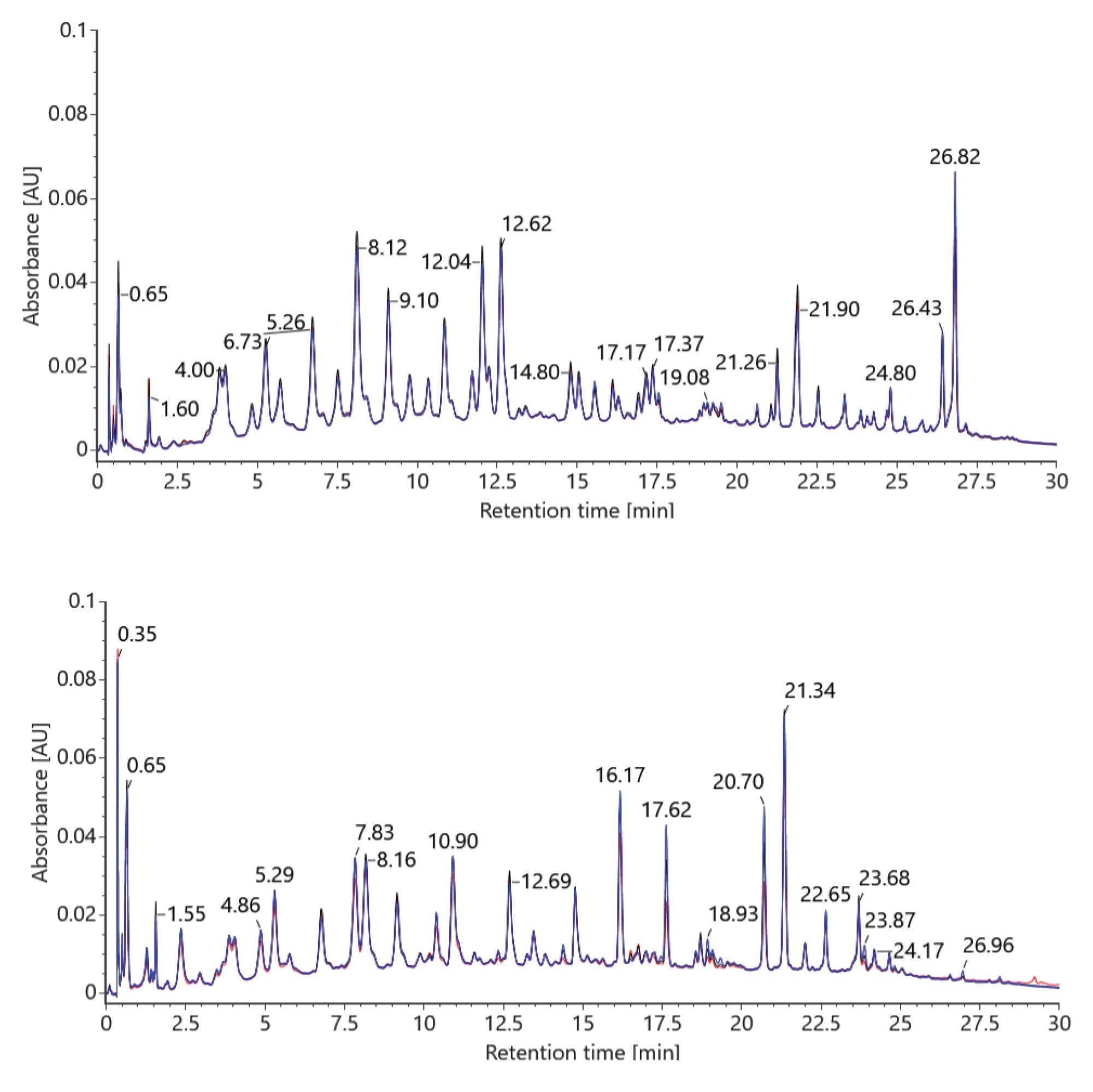
The first preparation includes a comparison of both manual and automated sample digestions yielding comparable chromatographic results for the RapiZyme MC1 and RapiZyme Cusativin. The same RapiZyme MC1 and Cusativin vial was split between the manual and automated procedure. The relative abundances of the RNA digestion products were found to be consistent between both sample preparation workflows with minor differences in a few select peak heights, as shown in Figure 3. It is theorized that these differences observed may be attributed to differences in the thermal ramp rate between the automated protocol, which ramped to temperature faster on the Peltier+, as compared to a slower ramp time on the manual protocol with a thermal cycler. The differences are described as minor for determining the sequence confirmation with the MAP Sequence app in waters_connect Software.
After the first round of testing to confirm equivalency between the manual and automated procedure, a set of eight samples were prepared by each enzymes protocol to investigate tube to tube reproducibility with the automated protocol. Three of the eight positions distributed on the PCR tube strip (A1, A5, and A8) were analyzed to evaluate the procedure in triplicate. Excellent repeatability was observed for both RapiZyme MC1 and Cusativin, demonstrating the pipetting precision of Andrew+ Robot as seen in Figure 4. The TUV data was used for the overlay to ensure a quantitative comparison between each replicate.
The GATA2 sgRNA sequence was inputted into mRNA Cleaver MicroApp and a list of predicted in silico cleavage products was generated for RapiZyme MC1 and Cusativin. The software was set to consider up to 4 missed cleavage sites for MC1 and 6 for Cusativin to account for partial digestion. The LC-MS data was processed in waters_connect MAP Sequence Application which automatically detect the presence or absence the RNA digestion products that were generated in silico in mRNA Cleaver.2 The MAP Sequence results can then be interpreted in the Coverage Viewer MicroApp to estimate the percentage of RNA sequence coverage for visualization of detected or non-detected with unique or ambiguous coverage (Figure 5). Both the manual and automated MC1 and Cusativin digestions achieved 100% coverage for GATA2 sgRNA.

Conclusion
RapiZyme MC1 and Cusativin offer analysts a unique approach to partial RNA digestion that produces longer overlapping digestion products and can be easily automated with high reproducibility that preserve valuable sequencing information for LC-MS analysis. In this study, we developed an automated RapiZyme RNase digestion procedure for processing up to eight samples in just 45 minutes using the Andrew+ Pipetting Robot and demonstrated achieving 100% sequence coverage of sgGATA2.
RapiZyme, Waters, Andrew+, QuanRecovery, MaxPeak, waters_connect, ACQUITY, BEH, IonHance, Xevo, and OneLab are trademarks of Waters Technologies Corporation. All other marks are the property of their respective owners.
References
- Addepalli, B., Johnston, T., Reidy, C., Lauber, M., Tunable Digestions of RNA Using RapiZyme™ RNases to Confirm Sequence and Map Modifications. Waters Application Note. 720008539, September 2024.
- Doneanu, C., Preston, C., Gorton, M., Johnson, T., Addepalli, B.,Yu, Y,. RNA Digestion Product Mapping Using an Integrated UPLC-MS and Informatics Workflow. Waters Application Note. 720008553, September 2024.
720008707, February 2025