For research use only. Not for use in diagnostic procedures.
In this application note, we introduce the workflow of TransOmics Informatics Software that has been specifically developed for the larger scale analysis of ion mobility MS data from proteomics and metabolomics data sets, comprising technical and biological replicates.
MSE and HDMSE are LC-MS-based acquisition strategies for life sciences that can be used for a variety of research focused applications and applied to the ACQUITY UPLC System and oa-Tof-based mass spectrometry platforms, including the SYNAPT G2-S HDMS System. In LC-HDMSE-based acquisitions, ions are separated in the gas phase by ion mobility and subsequently, the energy applied to the collision cell is alternated between a low and elevated state on a scan-to-scan basis, providing both qualitative and quantitative accurate mass precursor and product ion information that is retention and drift time aligned. The challenge is to make use of the high quality ion mobility resolved data that are generated. In this application note, we introduce the workflow of TransOmics Informatics Software that has been specifically developed for the larger scale analysis of ion mobility MS data from proteomics and metabolomics data sets, comprising technical and biological replicates.
Three replicates of each E.coli sample, differentially spiked with bovine serum albumin (BSA), alcohol dehydrogenase (ADH), Enolase and Glycogen phosphorylase B were analyzed. The injected on-column amounts for the spike protein in the first sample (Mixture 1) were 500 attomoles each and 4000, 500, 1000 and 250 attomoles for the second sample (Mixture 2), respectively. The peptides were separated and analyzed using a nanoACQUITY UPLC System coupled with a SYNAPT G2-S, operating at a mass resolution of >20k FWHM. The data were acquired in either LC-MSE or HDLC-MSE mode, an unbiased acquisition method in which the mass spectrometer switches between low and elevated energy on alternate scans with or without including ion mobility separation, respectively.
Online LC-MS instrumentation can generate considerable volumes of raw data, which often makes it impractical to run large numbers of biological and technical replicates. To overcome this, Nonlinear Dynamics developed an intelligent peak-modeling algorithm that can reduce data files by an order of magnitude. Using a wavelet-based approach, peaks are identified and peak models are created, and all relevant quantification and positional information is retained. TransOmics Informatics Software supports a number of common raw and processed data generated by LC-MS instruments.
The ability to combine data from multiple data independent analysis (DIA), UPLC-MSE or UPLC-HDMSE is required for comparative expression profiling studies. This enables the comparison of different experimental conditions or time-course studies using a high number of replicates. To combine and compare results from different runs, TransOmics Informatics Software aligns injections to compensate for between-run variations in the LC separation technique, shown in Figures 1 and 2. This results in increased reliability and reproducibility.
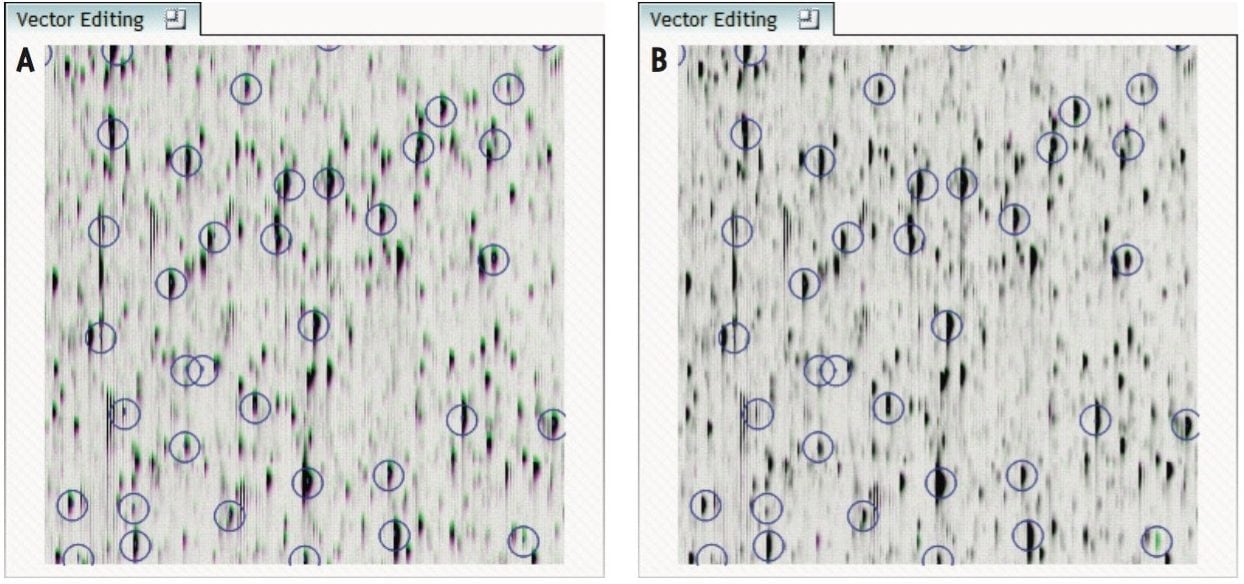
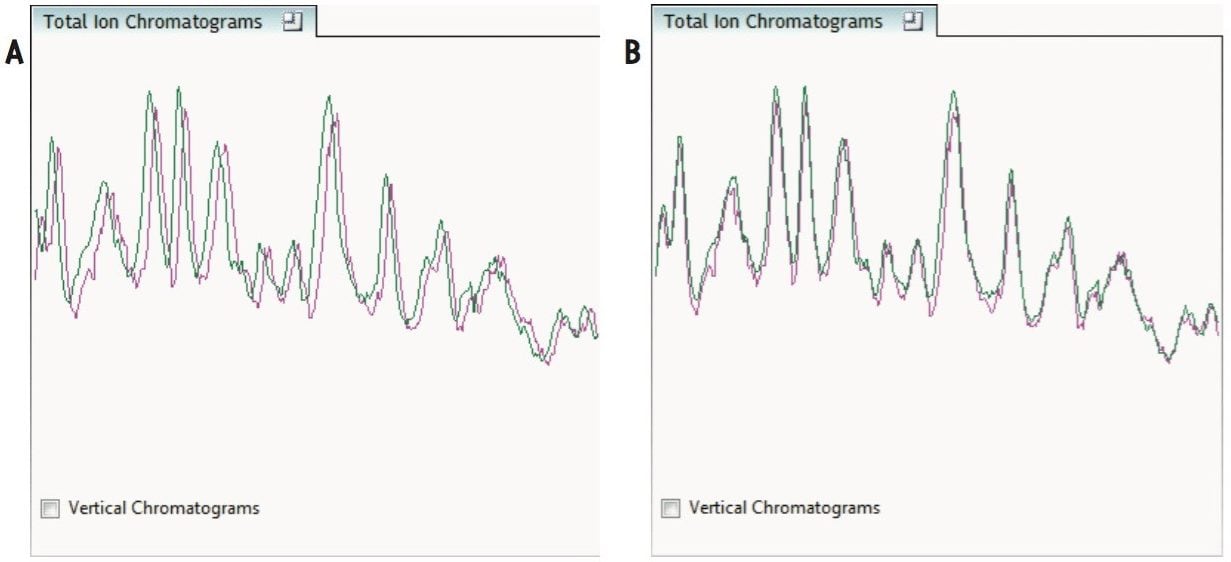
To ensure consistent peak picking and matching across all data files, an aggregate data set is created from the aligned runs. This data set contains all peak information from all sample files, allowing the detection of a single map of peptide ions. This map is applied to each sample, giving 100% matching of all chromatographic peaks with no missing values, enabling the application of multivariate statistical tools to explore data and measure differential analysis.
The peak picking algorithm handles complex samples and can discern overlapping isotopic peptide ion distributions. The end result is highly accurate detection, which saves time further down the workflow. The ion mobility separation of chromatographically co-eluting isotopic distributions is shown in Figure 3. It also generates complete datasets that contain no missing values, which allows obtaining reliable results from the multivariate statistical tools.
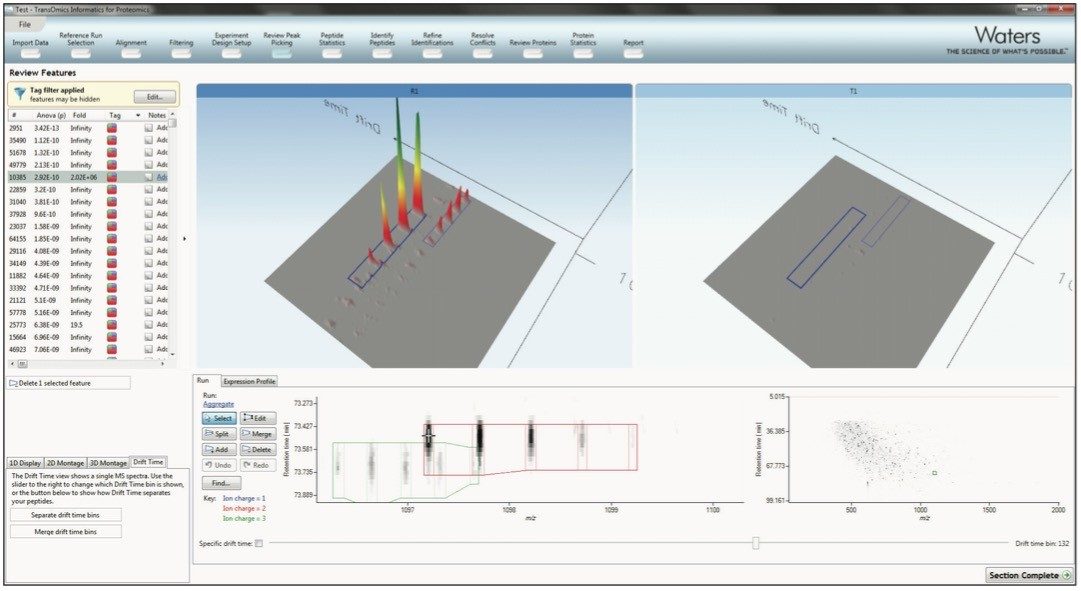
For the most accurate measurements, survey scan or low energy MSE data is used for quantitation. This significantly outperforms spectral counting, and allows for quantitation of peptide ions without (DIA) MSE or data directed analysis (DDA) MS/MS data. Additional fragmentation data can be gathered after the data analysis, using either a targeted DDA inclusion list or a non-targeted DIA experiment. After measuring each of the runs, the data are then normalized, so comparisons can be made between the runs.
Using advanced statistical tools – including ANOVA, power analysis, and q-values (for false discovery rate) – peptides can be user selected for identification.
MSE and HDMSE search results can be appended to the aggregate and quantitative results. Alternatively, once a list of peptide ions has been selected to be identified, the fragmentation data can be exported for searching using any of the search engines (e.g. PLGS, MASCOT, etc.) and databases supported.
For any features that do not have fragmentation data, a targeted inclusion list can be exported for a subsequent sample injection to gather further targeted MS/MS data, to increase the identification coverage for the peptides of interest.
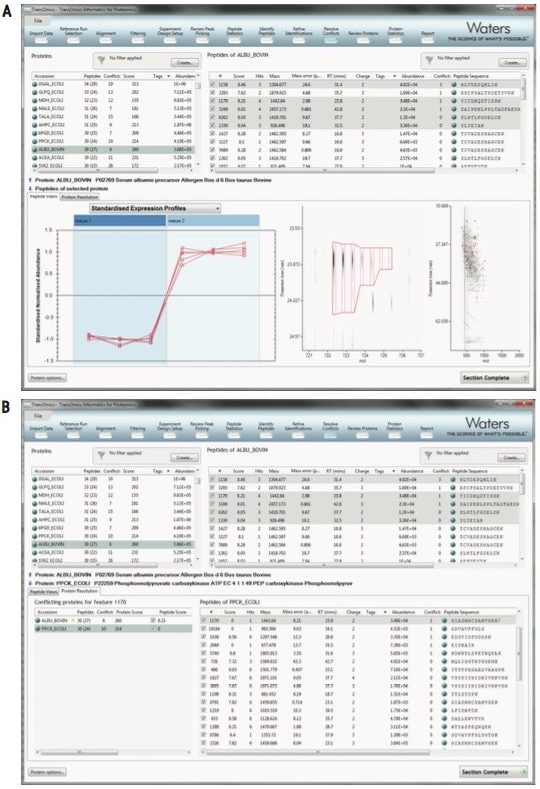
Quantitative data and qualitative fragmentation spectra search results are brought together to produce a peptide-based view of potentially interesting proteins in the experiment, with the ability to resolve conflicts when a peptide sequence is associated with more than one protein. It is easy to tag unique peptides and select the best ones as candidates for future MRM studies.
A final list of interesting proteins and their measurements, including a display of quantitative profiles in each group is automatically generated, as shown in Figure 4. Protein quantitation is based solely upon unique peptides. This provides reliable quantification for biological studies and publications.
Multivariate statistical analysis on protein measurements can be performed to make confident conclusions and generate a protein-based report of the experiment, as shown by the power analysis, dendrogram and expression examples in Figure 5. Protein measurements can also be exported to any custom bioinformatics workflow, and supplementary data or new results can be imported back into the analyzed experiment.

TransOmics Informatics Software has been specifically developed for large scale analysis of ion mobility MS data from proteomics and metabolomics.
720004344, May 2012