Tof MRM for the Quantification of Peptide Biomarkers in Human Glioblastoma with the Xevo™ MRT Mass Spectrometer
Matthew E. Dalya, Lee A. Gethingsa, Christopher J. Hughesa, Richard Locka, Nelofer Syedb
aWaters Corporation, Wilmslow, United Kingdom
bImperial College, London, United Kingdom
Published on October 08, 2025
Abstract
Benefits
- The Time-of-flight multiple reaction monitoring (Tof MRM) mode of acquisition on the Xevo MRT Mass Spectrometer demonstrates that high resolution mass spectrometry (HRMS) can be used for both quantitative and qualitative experiments
- The linear dynamic range and sensitivity of Tof MRM on the Xevo MRT Mass Spectrometer is comparable to MRM on the Xevo TQ Absolute Mass Spectrometer
- Statistical differences can be identified between marker peptides in treated and untreated glioblastoma cell lines using heavy labeled peptides for quantification
- The waters_connect™ Software Platform provides a complete workflow from data acquisition to data processing and results
Introduction
Glioblastoma is an aggressive form of brain cancer with few therapeutic options available, and correspondingly, the 5-year survival rate remains <10% and the median at 15 months.1 Of particular interest in disease progression is the activation of specific proteins via phosphorylation by protein kinases. Profiling these pathways through measurement of total activation, phosphorylation of specific residues and kinase abundance is essential when trialing of new therapies.2 Multi-reflecting time-of-flight (MRT) mass spectrometry (MS) with <1 ppm mass accuracy coupled with the selectivity of multiple reaction monitoring (Tof MRM3), allows for accurate and definitive characterization of post-translational modifications while also providing quantitative data surrounding the identified peptide markers. Further, through trapping and release of target ions timed with the pusher pulsing, the ion utilization of ions with specific m/z approaches 100%, termed “Enhanced Duty Cycle” (EDC), dramatically improving the sensitivity of the assay. Here, the use of Tof MRM using the Xevo MRT Mass Spectrometer is demonstrated for absolute quantification of phosphorylated peptides associated with glioblastoma.
Experimental
Human glioblastoma cancer cell lines (four in total) represented a control group and those treated with a metabolic therapy (ADI-PEG20).4 Cell lines were lysed in the presence of protease and phosphatase inhibitors and protein disulphide bonds were reduced with dithiothreitol (DTT) and alkylated with iodoacetamide prior to digestion using trypsin. Phosphopeptides were enriched using the Waters™ MassPREP™ Phosphopeptide Enrichment Kit (p/n: 186003864).
Additionally, quantitation curves were also generated using a HeLa digest spiked with the heavy labeled peptides (phosphorylated and non-phsophorylated). The heavy labeled peptides were constructed with the C-terminus lysine/arginine being labeled with 13C 15N.
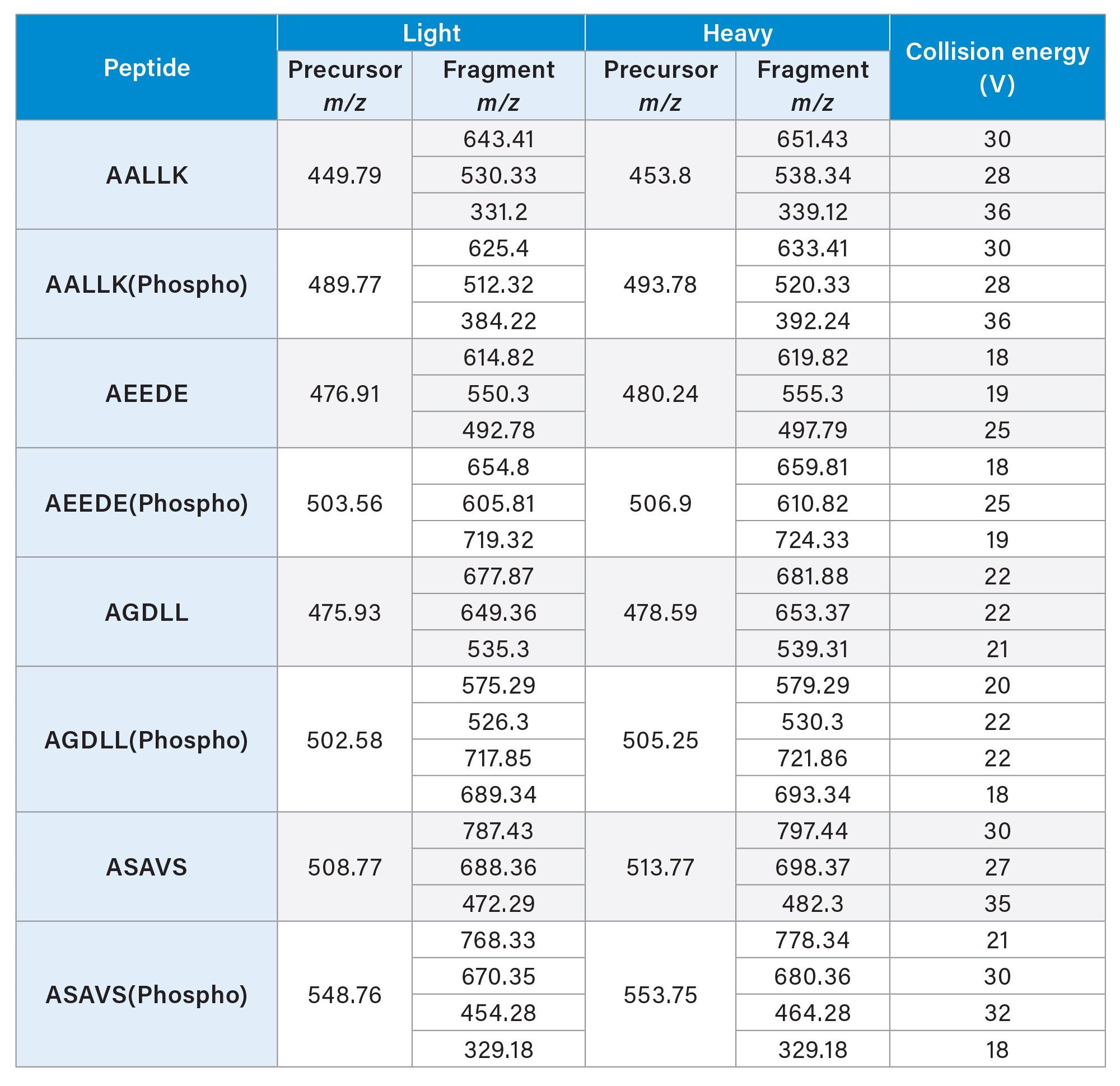
Digested samples or standards spiked with synthetic peptides were loaded onto Evotip™ Pure tips and separated using the Evosep™ One and the 30 SPD chromatographic method (48 minutes cycle time). The eluate was directed to a NanoLockSpray ion source with a 20µm fused silica emitter. Data were acquired on the Xevo MRT Mass Spectrometer, using the Tof MRM mode of acquisition monitoring three or four transitions per precursor with a dwell time of 70 ms, “Unit” precursor and product ion resolution (~1 Da) and collision energy optimized for each transition (Table 1).
The corresponding Xevo TQ Absolute Mass Spectrometer experiments consisted of using the same Evosep One inlet and chromatographic conditions with the NanoSpray ion source. The Xevo TQ Absolute Mass Spectrometer method consisted of the same precursor and product ion resolution as on the Xevo MRT Mass Spectrometer alongside the same dwell time per transition (70 ms).
In total, 52 transitions were monitored for both MS platforms. All data were subsequently processed using the waters_connect for Quantitation Application within the waters_connect Software Platform. Automatic integration of chromatographic peaks was completed by the software and a 1/x weighting applied, generating a linear calibration curve for each peptide. The RMS-1 option was used to generate the signal-to-noise ratio for each peak.
Results and Discussion
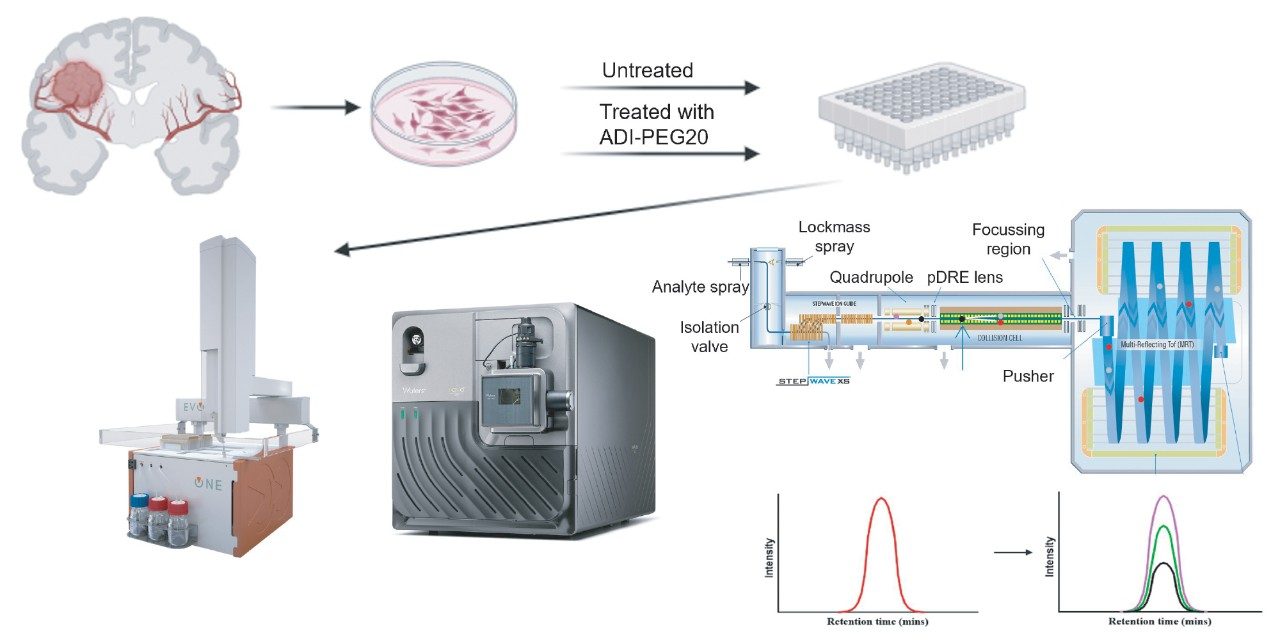
Discovery-based LC-MS experiments had been previously conducted to determine the peptide markers of interest, which could then be selected for absolute quantification (Product Solution p/n: 720008385). A total of four peptides were chosen and their synthetic equivalents (both native and phosphorylated) were generated as heavy analogues. Fresh glioblastoma cell lines (untreated and treated) were prepared to provide enriched phosphopeptides for LC-MS analysis, containing the heavy labeled synthetic peptides (Figure 1).
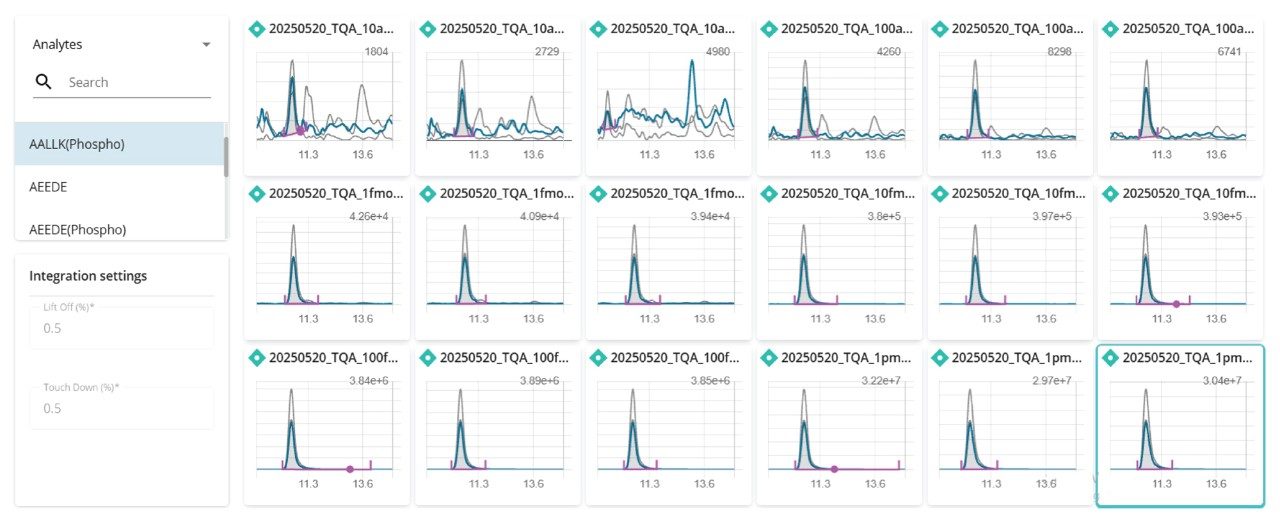
The subsequent data were processed using the MS Quan Application in the waters_connect Software Platform, highlighting the seamless workflow from method generation, acquisition, and import and processing, all achieved within the waters_connect Software Platform. Further, through the acquire and process functionality, data sets were automatically imported into the MS Quan Application after acquisition, increasing automative potential for this workflow. An example of the integrated quantifier ion peak throughout the calibration curve for peptide AALLK (Phospho) is shown in Figure 2.
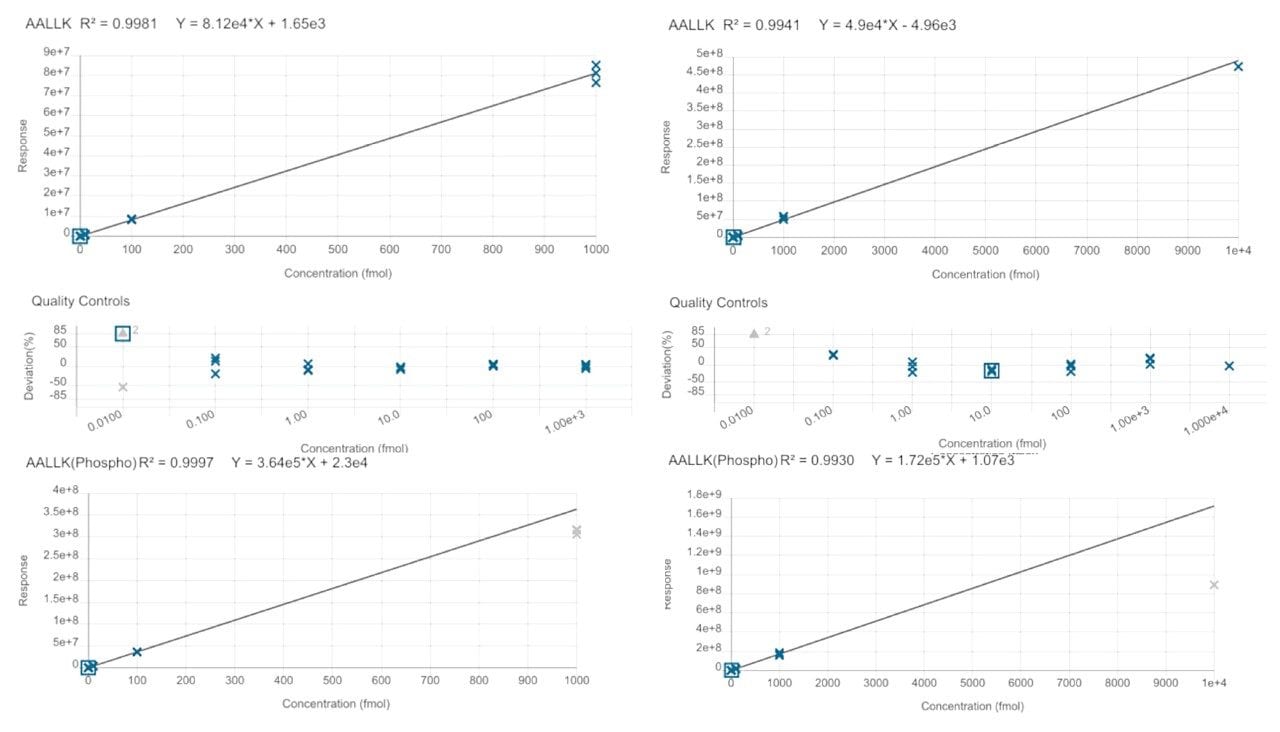
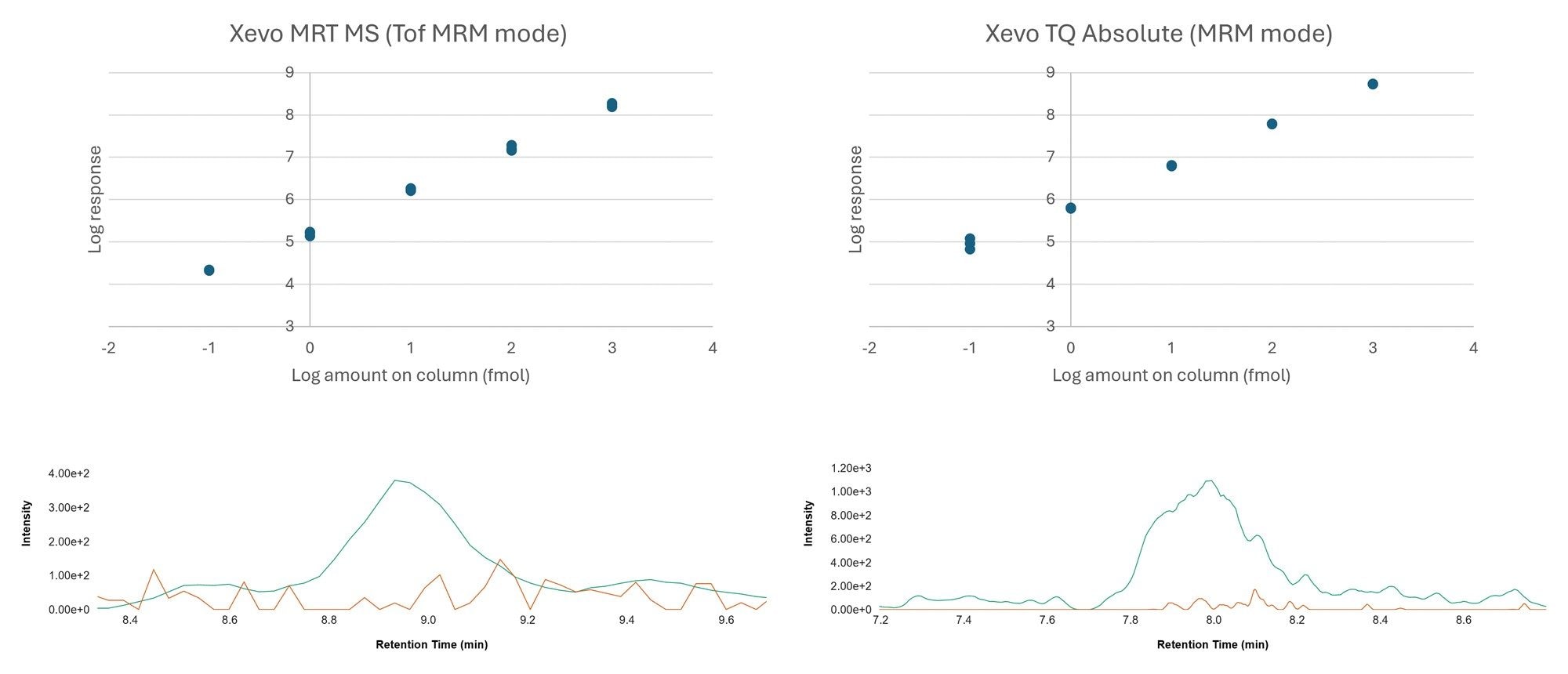
Calibration lines based on the HeLa digest spiked with the heavy labeled peptides, resulted in 4-orders of linearity for both MS platforms. Based on the peptide, AALLKASPK (non-phosphorylated and phosphorylated), this calibration curve is demonstrated with excellent linearity (R2>0.99) for all cases in Figure 3 (left panels generated from data acquired on the Xevo TQ Absolute MS and right panels from the Xevo MRT MS).
High-performing tandem quadrupoles mass spectrometers are renowned for their sensitivity and selectivity, granting them the place at the forefront of high-throughput targeted mass spectrometry, however lack the ability to retrospectively mine data for qualitative insights. Using Tof MRM, low levels of detection and quantitative accuracy have also been demonstrated on the Xevo MRT Mass Spectrometer. Combining this level of sensitivity with high resolution can be particularly advantageous with highly complex matrices where interference effects can become more problematic, such as the HeLa digest used in this study. In this study, "Unit” product resolution was to more appropriately compare the data acquired between the two instruments. Figure 4 provides an overview of the levels of sensitivity which can be achieved, yielding a LOD of 0.1 fmol (Xevo MRT Mass Spectrometer and Xevo TQ Absolute Mass Spectrometer). Comparatively assessing the ratios between both platforms regardless of sample loading highlights the similarity between Tof MRM and MRM experiments. Even at the lowest loading (10 amol), it was observed that both platforms provide a signal-to-noise ratio in the region of 6:1.
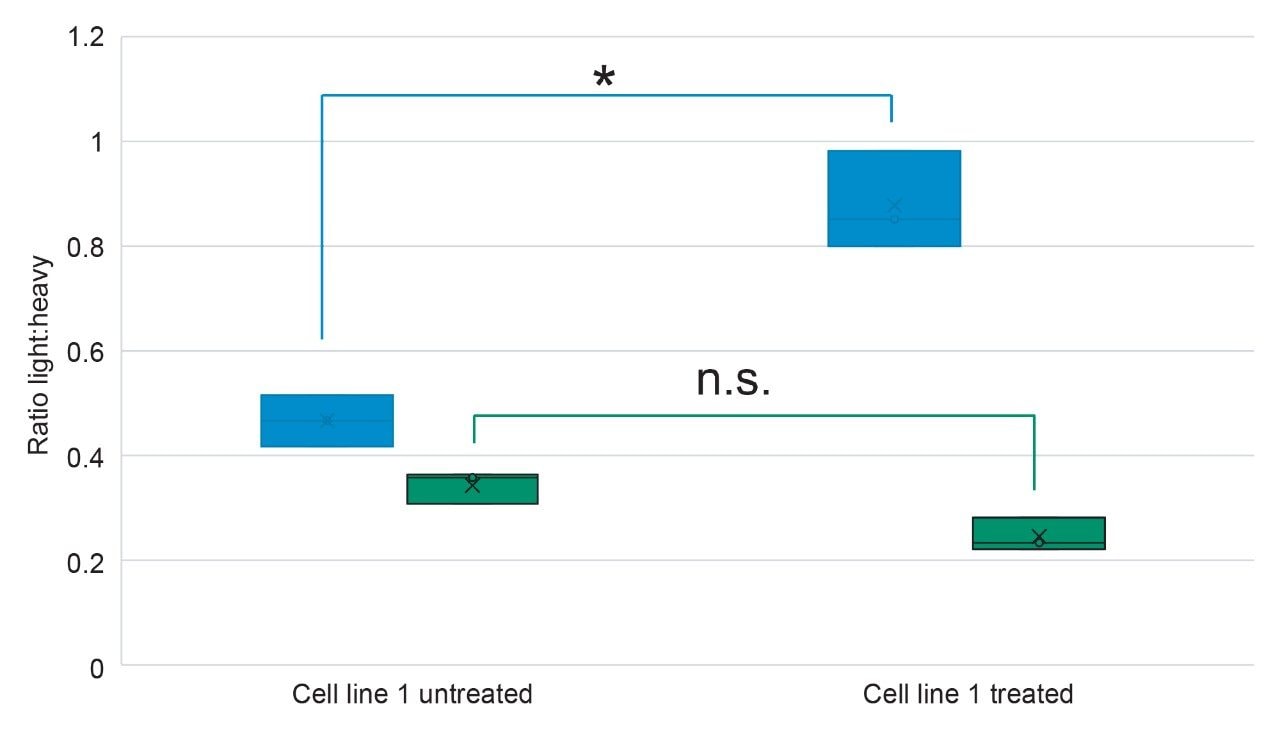
Measurements relating to the glioblastoma cell lines indicated that, although identified as potential marker peptides from previous DDA experiments, not all peptides provided a statistically significant difference (based on p-value) between untreated and treated when monitoring the light:heavy ratios (i.e., AGDLLpS), and were therefore unsuitable to use as markers of treatment with ADI-PEG20. However, absolute quantitation of a subset of peptide markers did show a significant difference between the cell lines, as is the case for AEEDEpS (p=0.0276) which corresponds to the calnexin (CANX) protein (Figure 5). CANX is involved in protein folding and quality control within the endoplasmic reticulum and is highly associated with glioblastoma development through the MEK/ERK/BNIOP3 pathway.5,6
Conclusion
The introduction of Tof MRM with EDC on the Xevo MRT Mass Spectrometer has elevated the system to a complete and verstaile Quan/Qual solution. In both cases, excellent linearity was achieved for the calibration curves generated (up to 4-orders of linearity) with excellent sensitivity and signal-to-noise ratio, providing LOD and LOQ’s of 0.1 and 1.0 fmol respectively. This methodology was then applied to a glioblastoma sample set, comprised of untreated and treated cell lines to highlight differentiating peptides following treatment with a metabolic therapy.
References
1. Poon, M.T.C., Sudlow, C.L.M., Figueroa, J.D. et al. Longer-term (≥ 2 years) survival in patients with glioblastoma in population-based studies pre- and post-2005: a systematic review and meta-analysis. Sci Rep 10, 11622 (2020). https://doi.org/10.1038/s41598-020-68011-4.
2. Qin, A., Musket, A., Musich, P. R., Schweitzer, J. B., Xie, Q. Receptor tyrosine kinases as druggable targets in glioblastoma: Do signaling pathways matter?, Neuro-Oncology Advances, Volume 3, Issue 1, January-December 2021, vdab133, https://doi.org/10.1093/noajnl/vdab133.
3. Tomcyzk, N., Wallace, A., Richardson, K., Grzyb, A., Wildgoose, J. Targeted High Resolution Quantification with Tof-MRM and HD-MRM, Application Brief, 720004728.
4. Hajji, N.; §Garcia-Revilla, J.; Soto, M. S.; Perryman, R.; Symington, J.; Quarles, C. C.; Healey, D. R.; Guo, Y.; Orta-Vazquez, M. L.; Mateos-Cordero, S.; et al. Arginine deprivation alters microglial polarity and synergizes with radiation to eradicate non-arginine-auxotrophic glioblastoma tumors. J Clin Invest 2022, 132 (6). DOI: 10.1172/JCI142137.
5. Wang et al. The dual role of calnexin on malignant progression and tumor microenvironment in glioma, Scientific Reports, 2024, 30796.
6. Liu et al. Calnexin promotes glioblastoma progression by inducing protective mitopagy through the MEK/ERK/BNIP3 pathway, Theranostics, 2025, 15(6), 2624–2648.
720008972, October 2025