In this application note, a rapid (few seconds), real-time method using DI in combination with post-ionization ion mobility separation to analyze lipidomic profiles in food and biological samples is presented.
The combination of real time desorption ionization and ion mobility MS offers a convenient solution for phenotypic identification and comparative lipidomic analysis.
Lipids are major constituents of food and biological tissues. Among lipid key properties are those to determine the caloric content, texture, and taste of food. Besides their importance in food and nutrition, lipid composition affects the physiology of living cells. Alterations in lipid profiles have been implicated in a wide range of pathologies in many types of organisms including plants and humans. Therefore, assessing lipid profiles and ratios between various lipid species can be indicative of the quality of food or health status of living organisms, as shown in Figure 1.
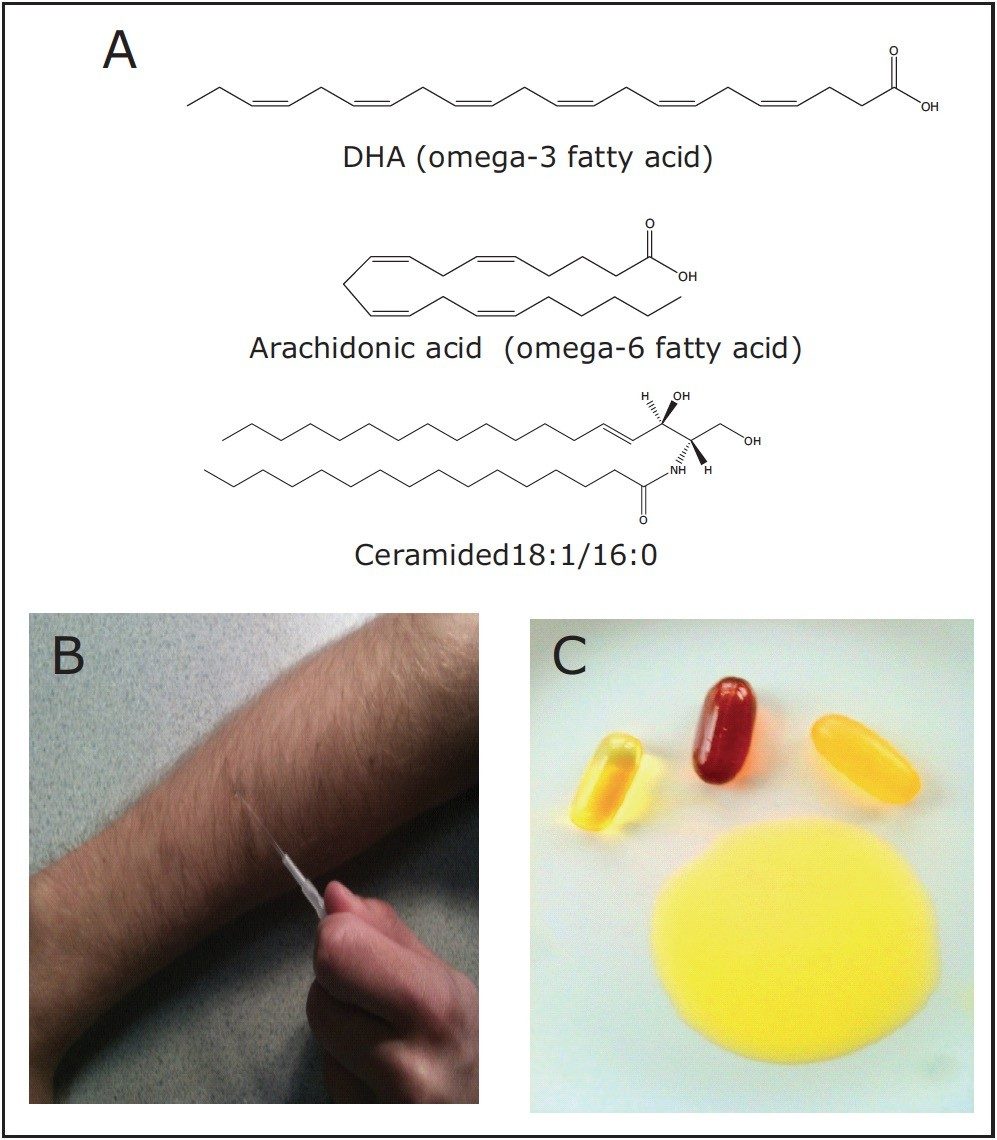
The analysis of lipid composition often requires very laborious and timeconsuming procedures. Furthermore, the detailed spatial distribution of lipid species on a surface is often missed using traditional sample preparation and lipid extraction protocols for large-scale lipid analysis (lipidomic analysis).
The use of desorption ionization (DI) techniques in lipidomics could provide a new level of description beyond the pure measure of lipid concentration. DI-MS techniques are useful for real-time, rapid, in-situ screening of various materials including food, plant, and animal tissue.1 In particular, DI-MS spectra of biological samples feature ions corresponding mainly to lipids. By molar quantities, the most abundant ionic molecular species in biological tissue, lipids ionize well under DI conditions.
The in-situ generation of a particular profile of lipid ions has been proposed for real-time molecular fingerprinting and diagnosis. Here, a rapid(few seconds), real-time method using DI in combination with post-ionization ion mobility separation to analyze lipidomic profiles in food and biological samples is presented.
No sample preparation is required. Samples were swiped on glass capillaries, which were held in the in metastable gas beam between the Direct Analysis in Real Time (DART, IonSense, MA, USA) ion source and SYNAPT G2 HDMS. Lipid standards and extracts were purchased from Avanti Polar Lipids (AL, USA). Edible oils were purchased at the local grocery store and blindly analyzed.
Chromatographic separation is not required. Analyses were conducted using a DART source coupled with a Waters SYNAPT G2 HDMS instrument. DART sources are designed to fit the Waters Xevo MS family of instruments. Acquisition time was 5 to 10 seconds.
|
Mass spectrometer: |
SYNAPT G2 HDMS |
|
Ionization: |
DART +ve and –ve |
|
Cone voltage: |
20 V |
|
Source temp.: |
120 °C |
|
DART temp.: |
50 to 450 °C |
|
Cone gas: |
30 L/h |
|
Desolvation gas: |
800 L/h (Nitrogen) |
|
IMS gas: |
90 mL/min (Nitrogen) |
|
IMS T-Wave velocity: |
833 m/s |
|
IMS T-Wave height: |
40 V |
|
Acquisition range: |
50 to 1200 |
For a rapid lipidomic analysis, we combined two emerging technologies: DART and ion mobility separation2 to analyze lipids extracted from biological samples.
Belonging to the DI techniques, DART is an atmospheric pressure ion source that instantaneously ionizes samples in open air under ambient conditions. DART employs an electrical discharge to create a plasma that produces helium metastables, which react with ambient water, oxygen, or other atmospheric components to produce charged water clusters. Protons are then transferred to the analytes.
Samples were swiped on glass capillaries, held in the in metastable gas beam between the DART ion source and SYNAPT G2 HDMS. Without the need for chromatographic separation, lipids were ionized by DART and guided into the mass spectrometer, where they traveled to the Ion Mobility Separation (IMS) cell. A T-Wave mobility separator used a repeating train of DC pulses to propel lipid ions through a nitrogen-filled IMS cell in a mobility dependent manner. Lipids migrated with characteristic mobility times (drift times) according to their size and shape before TOF detection, as shown in Figure 2.
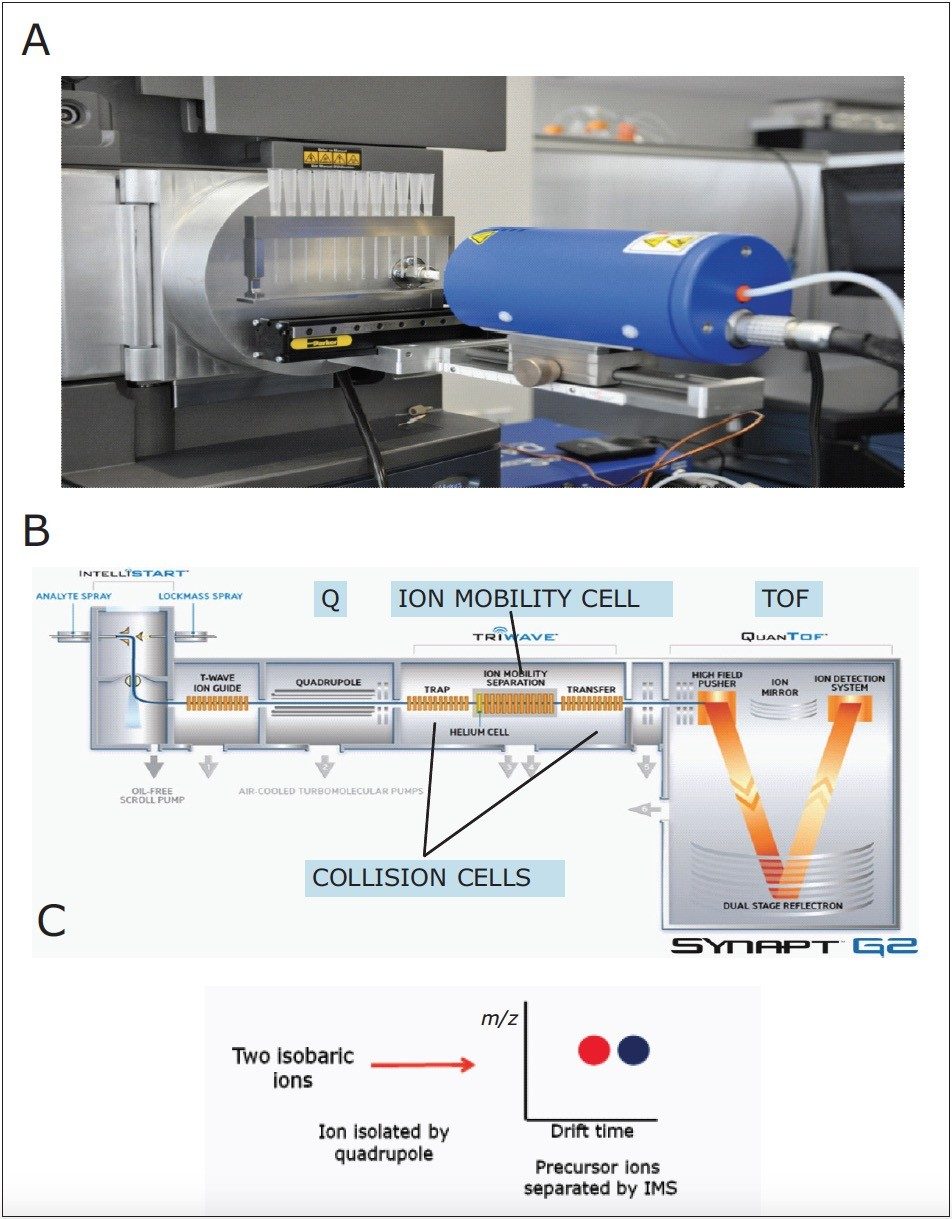
As an example of the power of such an approach, lipid profiles of edible oils (fish oil and olive oil), and lipids extracted from biological samples and human sebum, which is the oily matter that lubricates and waterproofs human skin, were analyzed, as shown in Figure 1. Lipid molecules with different acyl chain length or number of double bonds resulted in characteristic drift times. This enabled the separation and detection of key lipids, such as fatty acids and ceramides, on the millisecond time-scale without the need for prior derivatization or chromatography, as shown in Figures 1, 3, and 4. Ion mobility enabled the separation of the entire lipid profile of a sample on the millisecond time-scale, and a complete DART-IMS-TOF analysis required just a few seconds (0.1 min), as shown in Figures 3 and 4.
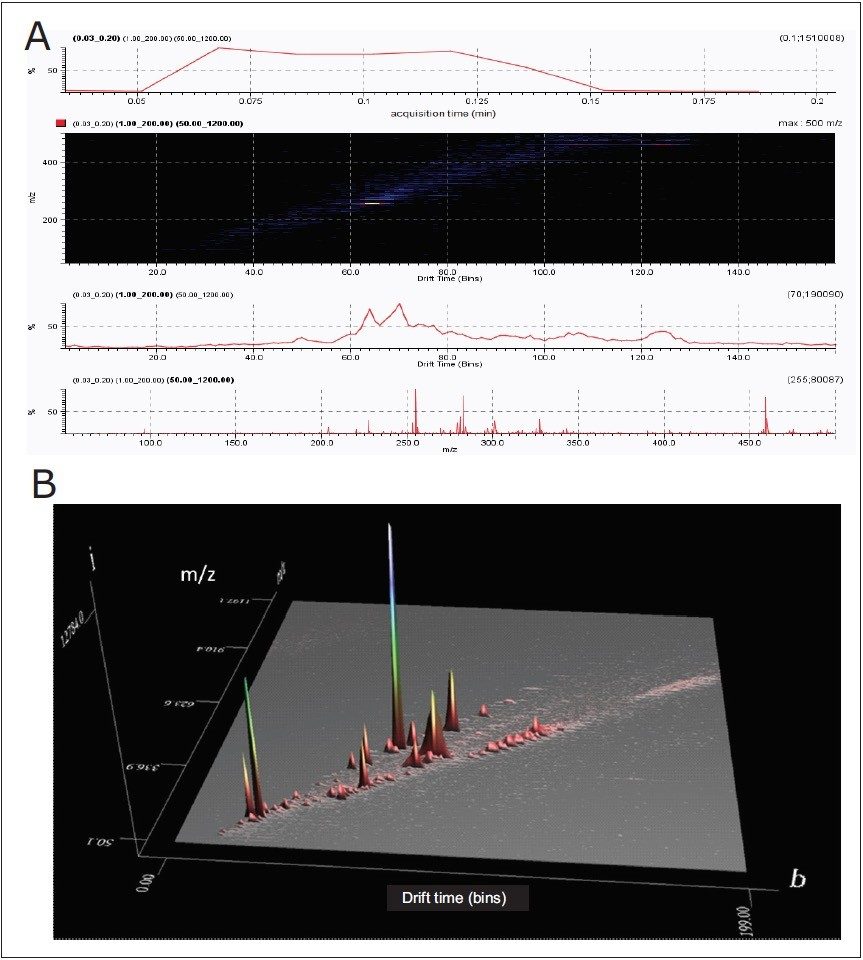
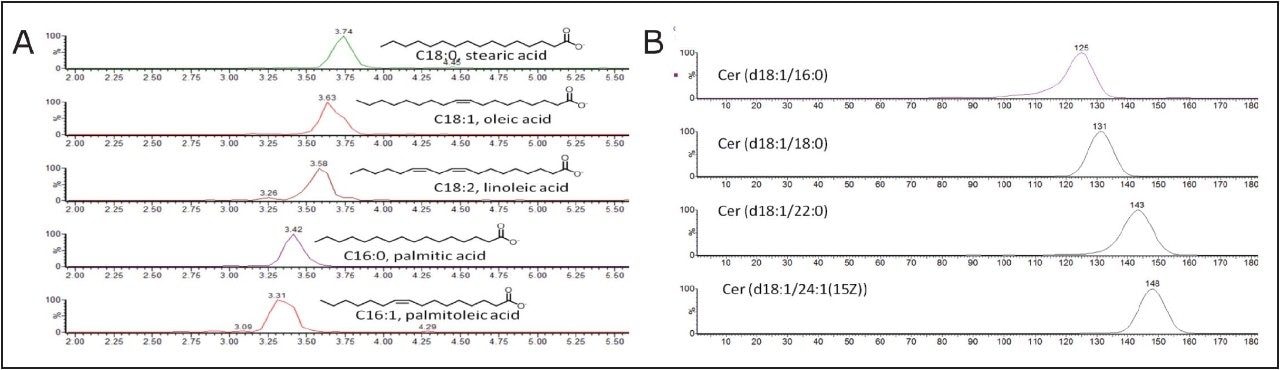
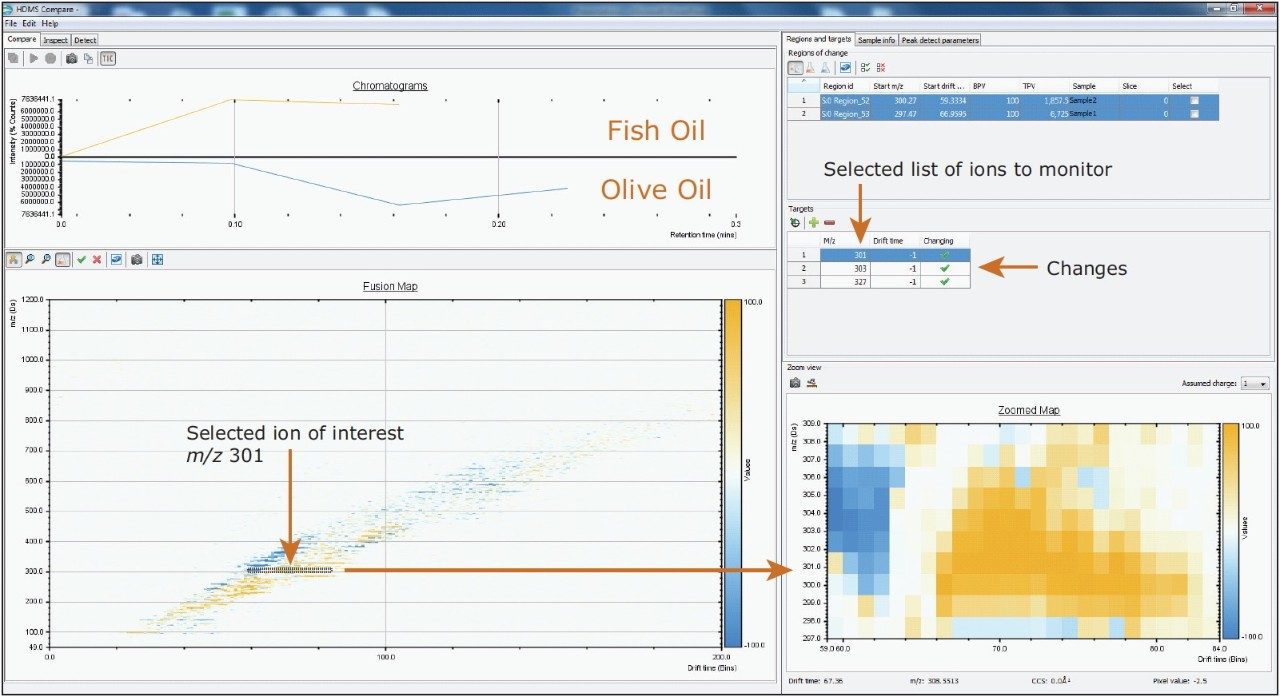
A comparison of lipid profiles of human sebum, shown in Figure 5, and edible oils shown in Figure 6 was done based on the separation capabilities of IMS-TOF/MS. HDMS Compare Software was used for a rapid binary comparison of different driftograms (masses versus drift time matrices). The drift time and spectral information associated with the components responsible for the differentiation can be extracted from the dataset and analyzed to better understand the underlying reasons for the observed differences.
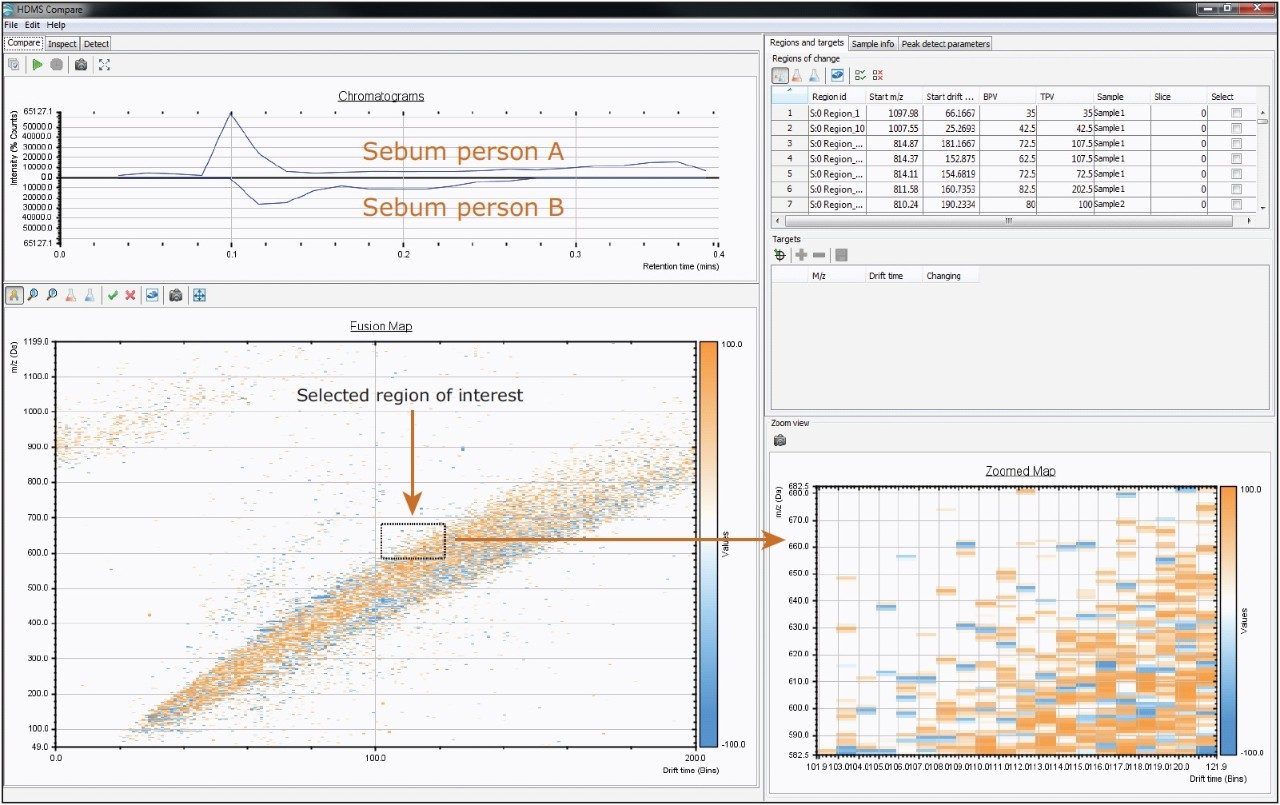
HDMS Compare Software also allows importing a list of target ions (mass and drift time) and reporting changes in the levels of these targets, as shown in Figure 6.
720004611, February 2013