One Pot, Mildly Acidic Digestion Protocols for Peptide Mapping Using Lys-C and RapiZyme™ Trypsin
Caitlin M. Hanna, Stephan M. Koza, Michael Zagieboylo, Steve Shiner
Waters Corporation, United States
Published on August 27, 2025
Abstract
Peptide mapping of therapeutic monoclonal antibodies (mAbs) is essential for confirming primary structure and assessing critical quality attributes (CQAs). However, the traditional sample preparation workflows can induce undesirable modifications, complicating downstream analysis. This study evaluates a streamlined, one-pot digestion protocol combining RapiZyme Trypsin and Mass Spectrometry (MS) Grade lysyl endopeptidase C (Lys-C) under low pH conditions that mitigate method-induced deamidation and oxidation.
Benefits
- Streamlined one-pot sample preparation for peptide mapping of biotherapeutic proteins
- Decreased levels of method-induced deamidation and oxidation achieved through the combined use of RapiZyme Trypsin and MS-Grade Lys-C in low pH digestion conditions
Introduction
Peptide mapping is used throughout the development and manufacturing lifecycle of therapeutic mAbs to deliver comprehensive information about primary structure and post-translation modifications. Sample preparation for peptide mapping is laborious and can result in undesired peptide modifications that complicate interpretation of certain CQAs. Asparagine deamidation and methionine oxidation, for example, can be induced under the alkaline conditions typically used in tryptic digestions.1 Upon analysis, these method-induced modifications are indecipherable from modifications that were present prior to digestion.
Low pH conditions can be used to mitigate method-induced deamidation and oxidation, but suboptimal trypsin activity under these conditions often leads to incomplete digestion. Combining trypsin with a secondary protease (e.g. Lys-C) has been shown to improve digestion completion at low pH.2 In this application note, RapiZyme Trypsin is combined with MS-grade Lys-C in a one pot, low pH digest of NISTmAb. We show that low pH decreases method-induced deamidation and oxidation and discuss the impact of pH and digestion time on digestion completion. Moreover, the one pot protocol eliminates the need for desalting prior to digestion, streamlining the sample preparation workflow.
Experimental
Sample Preparation
NISTmAb (NIST, p/n: 8671) and TCEP were added to solid GuHCl to prepare a 5 M GuHCl, 3 mM TCEP, and 5 mg/mL NISTmAb solution. Denaturation and reduction proceeded at room temperature for 30 minutes. IAM was added to a final concentration of 7 mM and the alkylation reaction proceeded in the dark at room temperature for 30 minutes followed by quenching with 3 mM TCEP. The denatured, alkylated, and reduced sample was diluted ten-fold with 50 mM histidine buffer (pH 5.5, 6.0, or 6.5) followed by addition of RapiZyme Trypsin (p/n: 186010108) at a 1:5 enzyme:protein (E:P) ratio and Lys-C (Marvelgent Biosciences, p/n: 14-LC-1MG) at a 1:50 E:P ratio. Digestion proceeded for 0.5, 1, or 3 hours at 37 °C and was quenched with 1% formic acid.
LC Conditions
|
LC system: |
ACQUITY™ Premier UPLC™ System |
|
Column: |
ACQUITY Premier Peptide CSH™ C18 Column, 130 Å, 1.7 µm, 2.1 x 100 mm (p/n: 186009488) |
|
Column temperature: |
60 °C |
|
Sample temperature: |
6 °C |
|
Injection volume: |
10 µL |
|
Mobile phase A: |
0.1% Formic Acid in H2O |
|
Mobile phase B: |
0.1% Formic Acid in ACN |
|
Detection λ: |
214 nm |
|
Sample vials: |
QuanRecovery™ MaxPeak™ 12 x 32 mm Propylene 300 µL Screw Cap Vials (p/n: 186009186) |
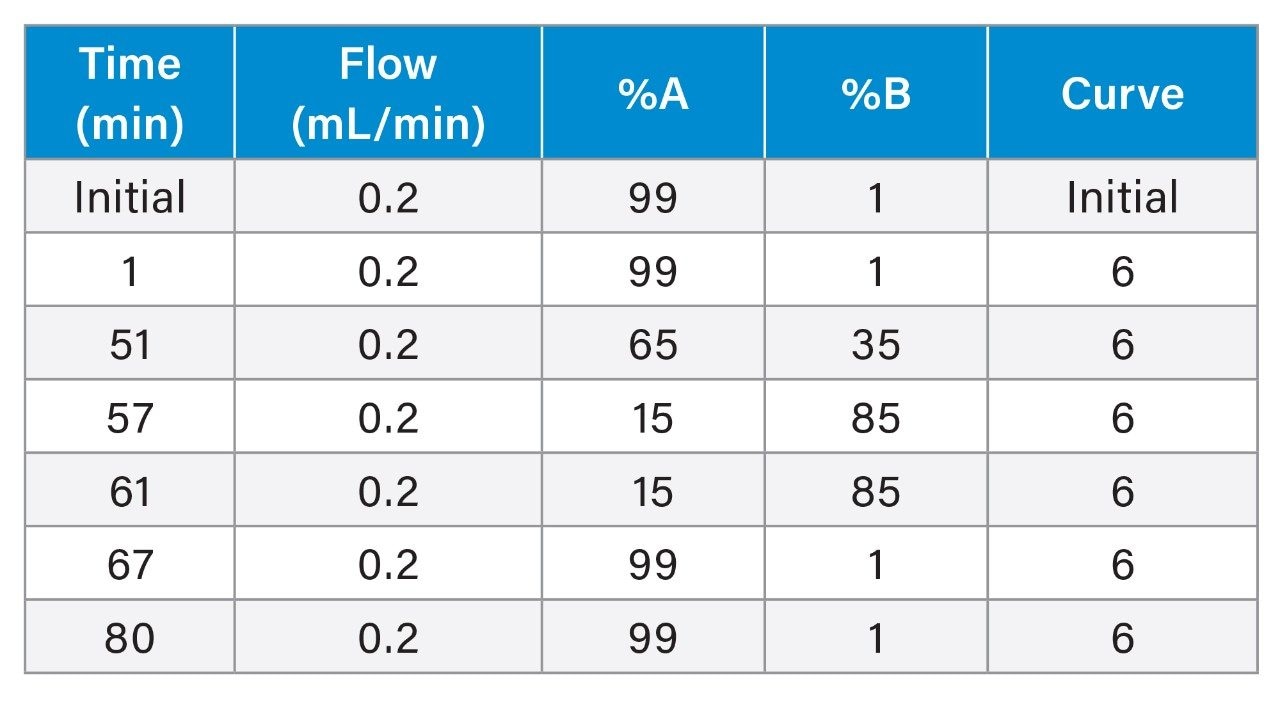
Gradient Table
Xevo™ G2-XS QTof Detector Settings
|
Mass range: |
100–4000 m/z |
|
Mode: |
ESI+ |
|
Sample rate: |
2 Hz |
|
Capillary voltage: |
2.2 kV |
|
Cone voltage: |
20 V |
|
Source temperature: |
120 °C |
|
Desolvation temperature: |
500 °C |
|
Cone gas flow: |
35 L/h |
|
Desolvation gas flow: |
600 L/h |
|
Data management: |
waters_connect™ Software |
Results and Discussion
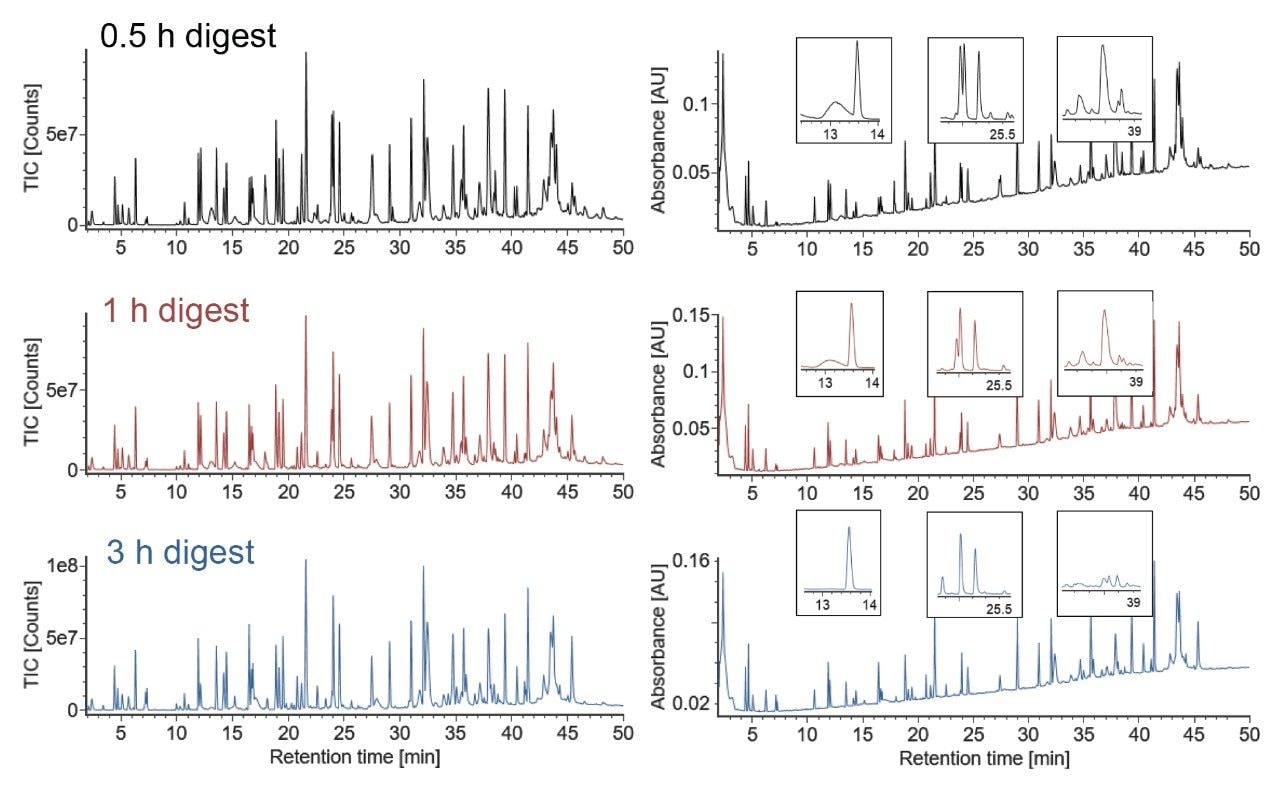
One pot, combined Lys-C/RapiZyme Trypsin digests of NISTmAb were prepared at pH 5.5, 6.0, and 6.5. Prior to digestion, NISTmAb was denatured with GuHCl, reduced with TCEP, and alkylated with IAM. The samples were diluted ten-fold prior to digestion to decrease the concentration of GuHCl, a known trypsin inhibitor, eliminating the need for a desalting step. Three digestion times were assessed at each pH (0.5, 1, and 3 hours). Representative chromatograms of pH 6.0 digests are shown in Figure 1. The chromatographic profile of the digest changes with increased digestion time; selected areas of significant change are shown in the zoomed insets. Similar time-dependent changes are observed at pH 5.5 and pH 6.5.
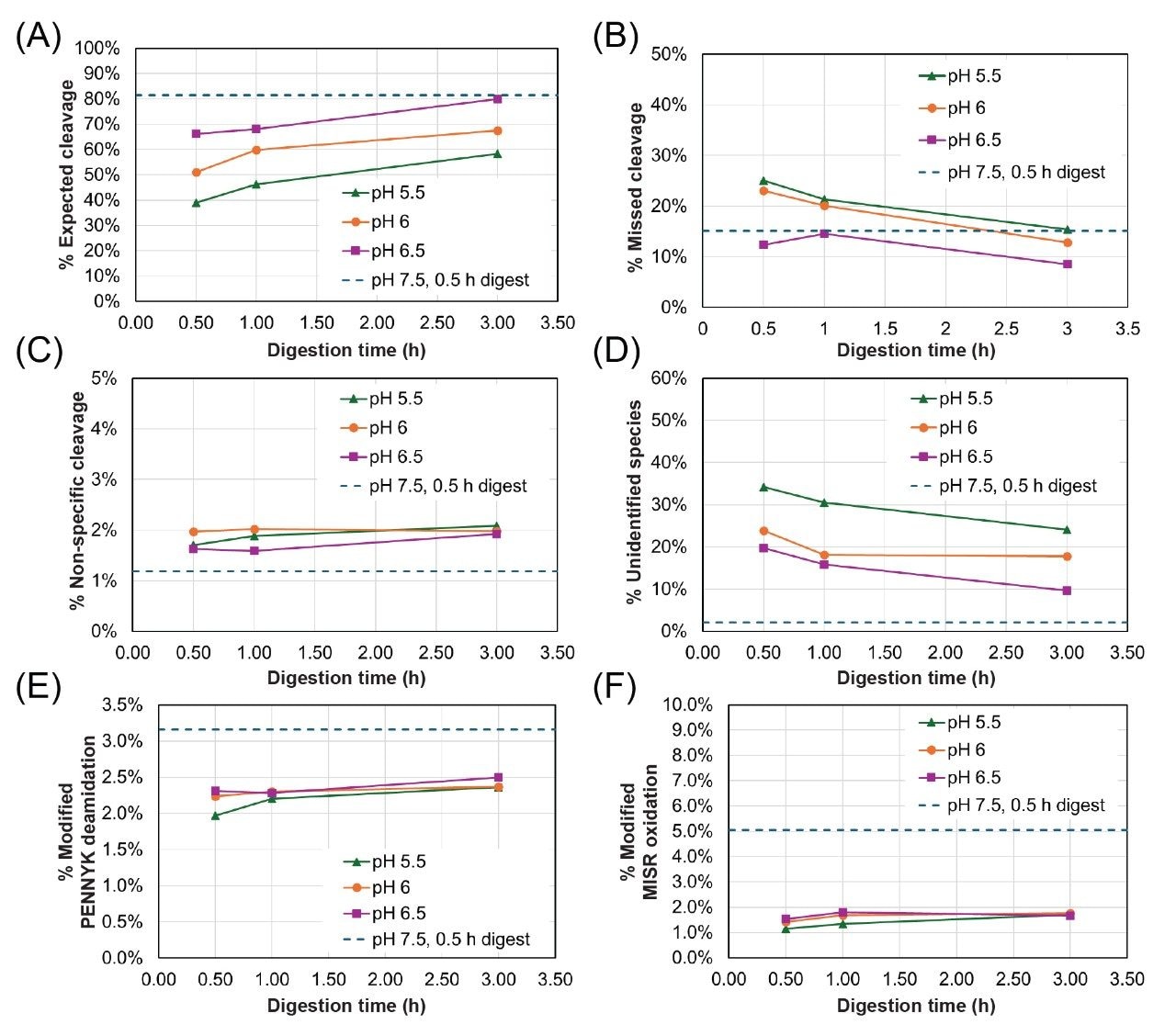
To illustrate the impacts of pH and incubation time on digestion completion, the relative abundance of expected cleavages, missed cleavages, non-specific cleavages, and unidentified species as a function of digestion time for all pH conditions are shown in Figure 2 (A-D). Relative abundances were calculated using the total MS response of the target cleavage type relative to the total response of all NISTmAb-derived peptides. The dashed traces in all plots in Figure 2 reflect a benchmark value from a one pot, 0.5 hour Lys-C/RapiZyme Trypsin digest of NISTmAb at pH 7.5.
The abundance of expected cleavages increases as a function of both pH and digestion time (Figure 2A). A 3 hour digest at pH 6.5 achieves a comparable level of expected cleavages to the benchmark 0.5 hour digest at pH 7.5. The abundance of missed cleavages, on the other hand, decreases as a function of pH and digestion time (Figure 2B). This improvement with increasing pH is expected given that the optimal pH range for both trypsin and Lys-C is mildly alkaline (pH 7–9). Under all one-pot conditions, the relative abundance of missed cleavages is high relative to workflows that include a desalting step (e.g., Waters™ PeptideWorks™ Tryptic Protein Digestion Kit) due to the low concentration of GuHCl present in the one-pot digests.4 Non-specific cleavages show a slight increase with digestion time but a dependence on pH is not clear based on the presented data. Regardless, the non-specific cleavages are low (<2.5%) across all tested conditions. A high abundance of unidentified species is observed under all reported low pH conditions; MS1 data suggests that these species are NISTmAb peptides containing both missed and non-specific cleavages, but identification could not be confirmed with available MSE data. The abundance of these unidentified species decreases with both increasing pH and increasing incubation time.
Method-induced deamidation and oxidation were assessed by quantifying the abundance of the deamidated “PENNYK” peptide (T37, GFYPSDIAVEWESNGQPENNYK) and the oxidized “MISR” peptide (T21, DTLMISR). The abundance of modified PENNYK and MISR peptides under all digest conditions are shown in Figure 2E-F. A slight increase in PENNYK deamidation and MISR oxidation is observed with increasing incubation time but the relative abundance of these modified peptides is lower than the benchmark 0.5 hour, pH 7.5 digest under all low pH conditions, regardless of incubation time.
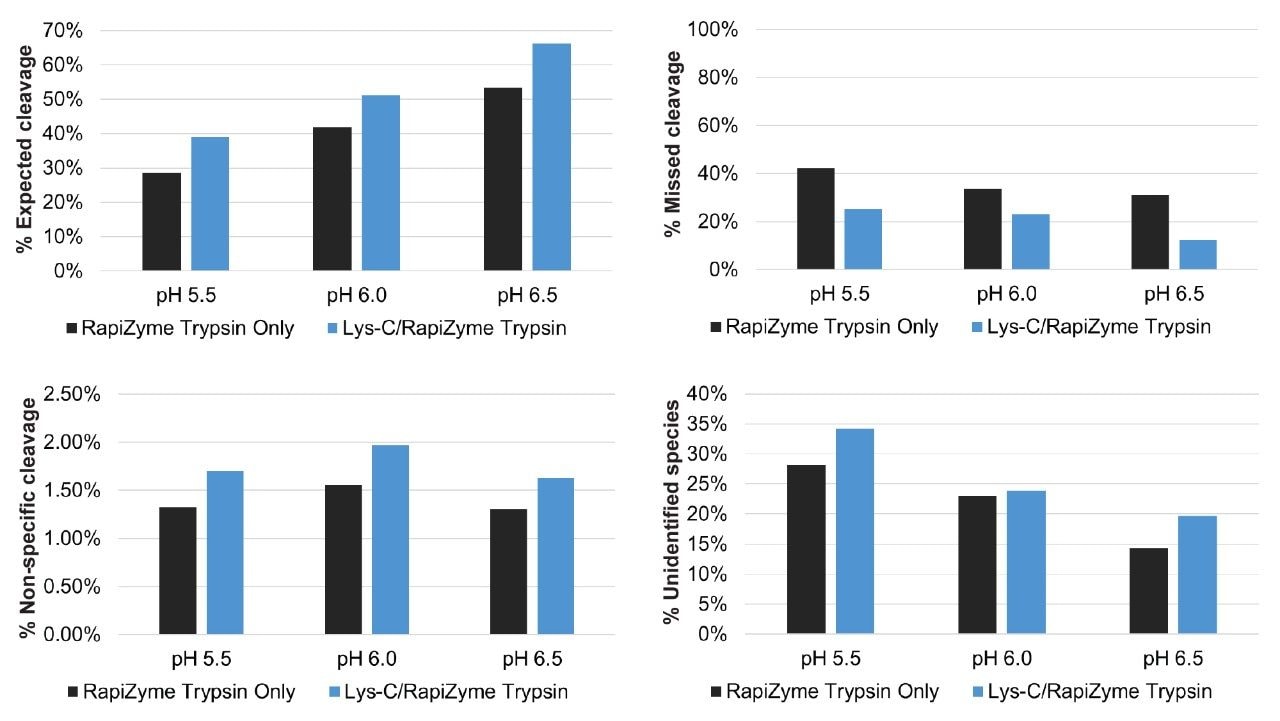
To highlight the benefit of combining Lys-C with RapiZyme Trypsin, the relative abundance of expected cleavages, missed cleavages, non-specific cleavages, and unidentified species in one pot, 0.5 hour RapiZyme Trypsin and combined Lys-C/RapiZyme Trypsin digests of NISTmAb are shown in Figure 3. The addition of Lys-C increases the abundance of expected cleavages and decreases the abundance of missed cleavages. Thus, the addition of Lys-C improves digestion completion under all low pH conditions. It should be noted that the abundance of non-specific cleavages and unidentified species increases slightly with the addition of Lys-C under the reported low pH conditions.
RapiZyme Trypsin is homogeneously methylated, rendering it resistant to autolysis even at high enzyme:protein ratios.3 To assess autolysis in combined RapiZyme Trypsin and Lys-C digests, a 0.5 hour blank digest was performed under optimal pH conditions (pH 7.5, no GuHCl). The total ion chromatograms (TICs) of blank Lys-C/RapiZyme Trypsin and blank RapiZyme Trypsin digests are shown in Figure 4. Peaks identified as trypsin or Lys-C autolysis species are annotated with the corresponding protease and sequence start and end residue number. Unknown peaks are annotated with their observed mass. Peaks annotated in bold are present only in the Lys-C/RapiZyme Trypsin digest. One Lys-C autolysis species and two unidentified species (1973.0 and 1366.7 Da) that are not observed in the RapiZyme Trypsin digest are observed in the combined digest. Lys-C remains largely intact under mock-digestion conditions. Extracted ion chromatograms (XICs) for two autolytic peptides were generated for all reported NISTmAb digests; the corresponding peak areas are plotted as a function of digestion time in the bottom of Figure 4. As expected, both autolytic peptides increase in abundance as a function of digestion time. Higher pH conditions yield a greater abundance of the trypsin 46–49 peptide while lower pH conditions yield a greater abundance of the Lys-C 158–204 peptide.
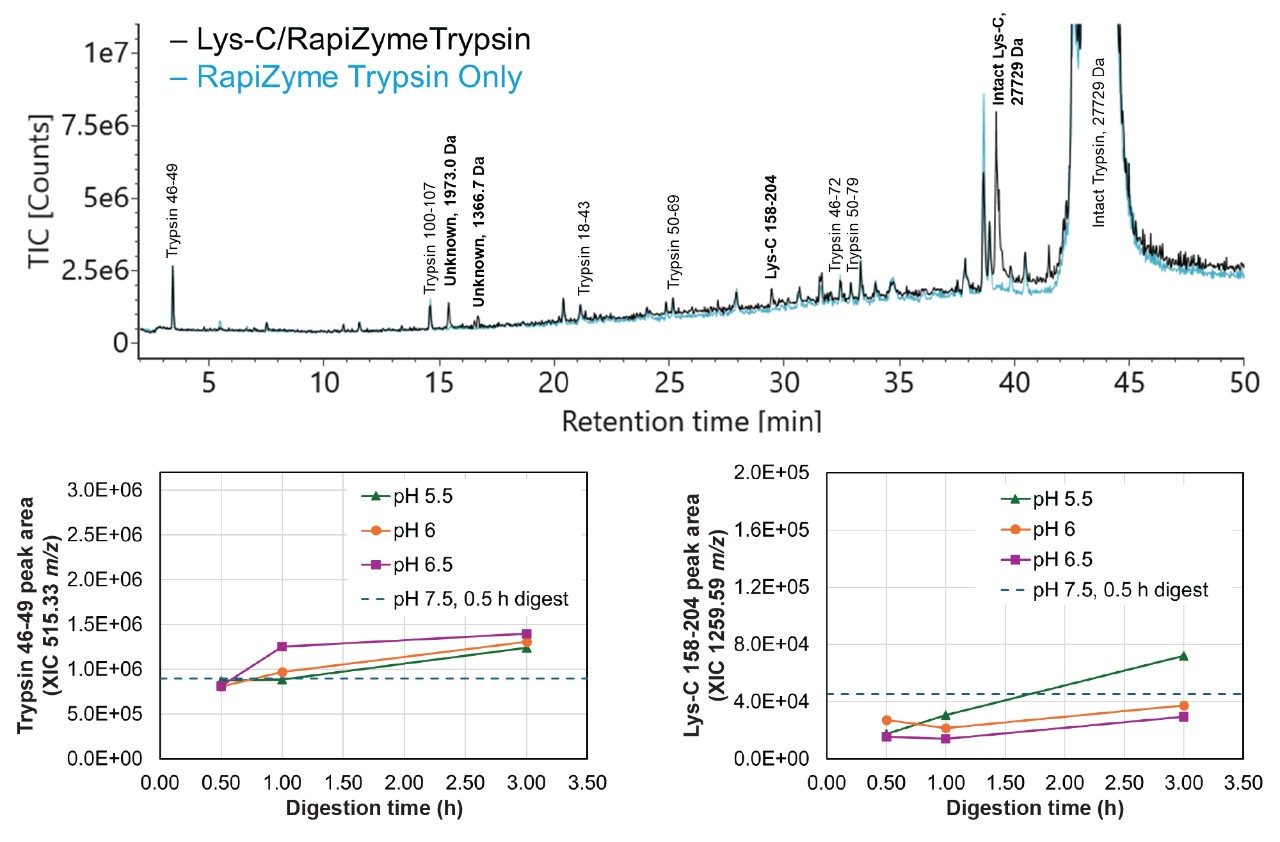
Figure 4. TIC of 0.5 hour blank digests performed at pH 7.5 without GuHCl. The black trace corresponds to a Lys-C/RapiZyme Trypsin blank digest (mock 1:5 trypsin:protein, 1:50 Lys-C:protein ratios) and the blue trace corresponds to a RapiZyme Trypsin-only digest (mock 1:5 trypsin:protein ratio). Identified autolysis peaks are annotated with the protease name and sequence start and end point. Unknown peaks are annotated with the highest abundance m/z. Peaks annotated in bold are present only in the Lys-C/RapiZyme Trypsin digest. Peak areas for the trypsin 46–49 and Lys-C 158–204 autolytic peptide XICs from the low pH and benchmark digests are shown in the bottom plots.
Conclusion
The combined use of RapiZyme Trypsin and MS-grade Lys-C in a low pH, one pot digestion protocol offers an efficient approach to sample preparation for peptide mapping of mAbs. We show that the pH can be tuned to minimize method-induced peptide modifications and discuss the impact of pH and incubation time on digestion completion. Blank digests under optimal conditions highlight the autolysis resistance of RapiZyme Trypsin even when used in combination with Lys-C. This methodology delivers high-quality peptide mapping results while simplifying sample preparation. It is expected that these protocols can also be amenable to focused peptide mapping studies.
References
- Chelius D, Rehder DS, Bondarenko PV. Identification and Characterization of Deamidation Sites in the Conserved Regions of Human Immunoglobulin Gamma Antibodies. Anal. Chem. 2005; 77, 6004.
- Cao, M. et al. An Automated and Qualified Platform Method for Site-Specific Succinimide and Deamidation Quantitation Using Low-pH Peptide Mapping. J. Pharm. Sci. 2019; 108, 3540–3549).
- Ippoliti, S., Zampa, N., Yu, Y. Q., Lauber, M. A. Versatile and Rapid Digestion Protocols for Biopharmaceutical Characterization Using RapiZyme™ Trypsin. Waters Application Note, 720007840EN. January, 2023.
- Hanna, C. M., Danaceau, J. P., Koza, S. M., Shiner, S., Trudeau, M. Quick and Robust Sample Preparation for Tryptic Peptide Mapping With the PeptideWorks™ Kit using Simple, Automatable Workflows. Waters Application Note, 720008019EN. July, 2023.
Featured Products
720008999, August 2025