Automated Screening for Pharma Process Development Workflows with the Waters ACQUITY™ RDa™ Detector, RemoteAnalyzer® and AnalyzerPro® XD
Christopher Henrya, Michael Jonesa, Scott J Campbellb
a Waters Corporation, United States
b SpectralWorks, United Kingdom
Published on November 14, 2025
Abstract
In this application note, a quick, simple sample submission for the acquisition of accurate mass measurements is demonstrated using the ACQUITY RDa Detector. Targeted and untargeted screening workflows are easily accessible to synthetic and process chemists to accelerate process optimization during drug development, with fully-integrated walk-up software RemoteAnalyzer and the complementary mass spectrometry processing software AnalyzerPro XD.
Benefits
- Routinely access untargeted accurate mass measurements for rapid assessment on API (Active Pharmaceutical Ingredient) and impurity profile during synthetic process design and evaluation.
- Inclusion of multiple ion chromatograms (EICs) for targeted analysis.
- Single, simple, and intuitive sample submission interface for rapid sample throughput.
- Completed results can be emailed or exported to an ELN (electronic notebook).
- Compatible with all waters_connect™ and MassLynx™ instruments.
Introduction
Scaling discovery chemistry synthetic routes to pilot plant and manufacturing scale is an integral component of the drug development pipeline. Understanding optimum reaction conditions is key to maximizing the yields of desired products. Additionally, this simultaneously reduces the production of inadvertent side products, which can lower yields, may be harmful, and can require additional separation steps.1
The need for continuous productivity improvements in drug development has led to an increased reliance on open access software2 to facilitate a simplified sample submission process, and the receipt of condensed, easily digestible results for rapid decision making.
Continuous improvements in mass spectrometry (MS) and interface technologies, combined with advanced liquid chromatographic (LC) techniques for high-throughput qualitative and quantitative analysis, have resulted in a wider scope of applications in the pharmaceutical field. LC-MS tools are increasingly used to analyze pharmaceuticals across a variety of stages during discovery and development.3
Increased confidence through compound characterization is possible through the accurate mass measurements provided, for example, by Time of Flight (ToF) High Resolution Mass Spectrometry (HRMS). The implementation of HRMS into these environments has often been difficult due to perceived complexity of instrumentation and data interpretation.
With the ACQUITY RDa HRMS Mass Detector and its automatic, push-button setup, accurate mass data is now accessible without the need for HRMS expertise. Combined with the open access software RemoteAnalyzer (SpectralWorks Ltd., Runcorn, UK), chemist end-users now have a simple, quick sample submission interface to accelerate the drug development process using both targeted and untargeted analysis enabling efficient, high-yield synthesis pathways.
To demonstrate the benefits of this platform, a sample of the anti-diabetic drug ‘glipizide’ was degraded under acidic, basic, and oxidative conditions to simulate the generation of different impurities in process optimization setting.
Using the “Peak Detection” untargeted workflow within RemoteAnalyzer, a specified number of ‘top intensity’ peaks above a defined threshold can be detected and an accurate mass returned. A targeted dimension was also included - in this case, a requested extracted ion chromatogram (EIC) of the parent compound glipizide (1-cyclohexyl-3-[[p-[2-(5-methylpyrazincarboxamido) ethyl] phenyl] sulfonyl] urea: m/z 446.18565) - as a mass confirmation experiment.
Acquired data were then transferred to the complementary MS processing software AnalyzerPro XD, where it was processed and screened against mol. Files of known glipizide degradants4 and further supported by online library searching (PubChem).
Experimental
Sample Description
Samples of glipizide were prepared and analyzed as per the application note 720007510.4
Data Management
|
MS software: |
waters_connect 4.0.0 |
|
Informatics: |
RemoteAnalyzer® v4.61.4678.0, AnalyzerPro XD® v1.16.93339.39575, (SpectralWorks Ltd. Runcorn, UK) |
Results and Discussion
When generating data on the waters_connect Platform using RemoteAnalyzer as the user interface software, samples can be submitted individually, as a batch (manually or through a csv. file) or as a 96-well plate using a csv. file. For this experiment, samples were submitted manually in vials.
On submission of the samples, an email was received to inform the operator that a batch had been submitted. On completion of each sample, using the acid degradation as an example, a PDF containing peak detected chromatogram (Figure 1A), an EIC for glipizide (Figure 1B) and mass spectra associated with each detected peak (Figure 1C) were published and emailed.
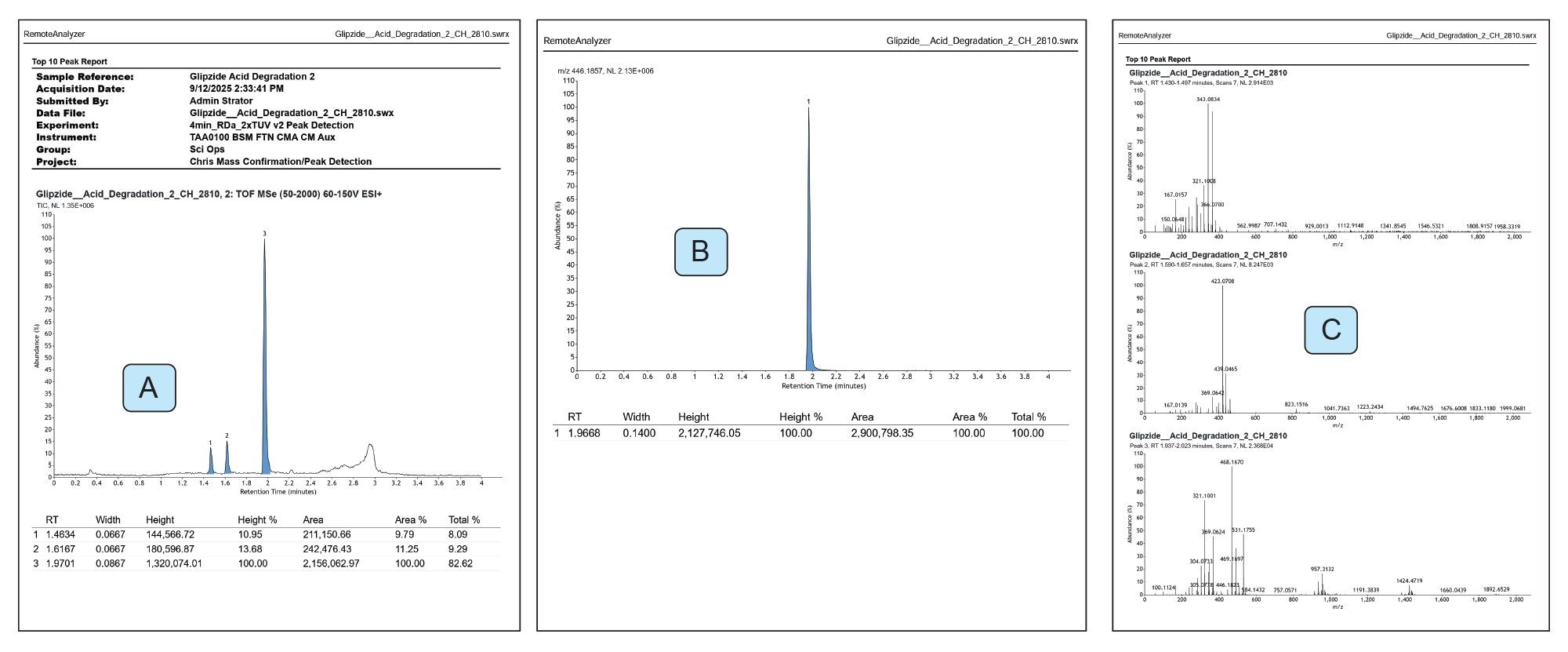
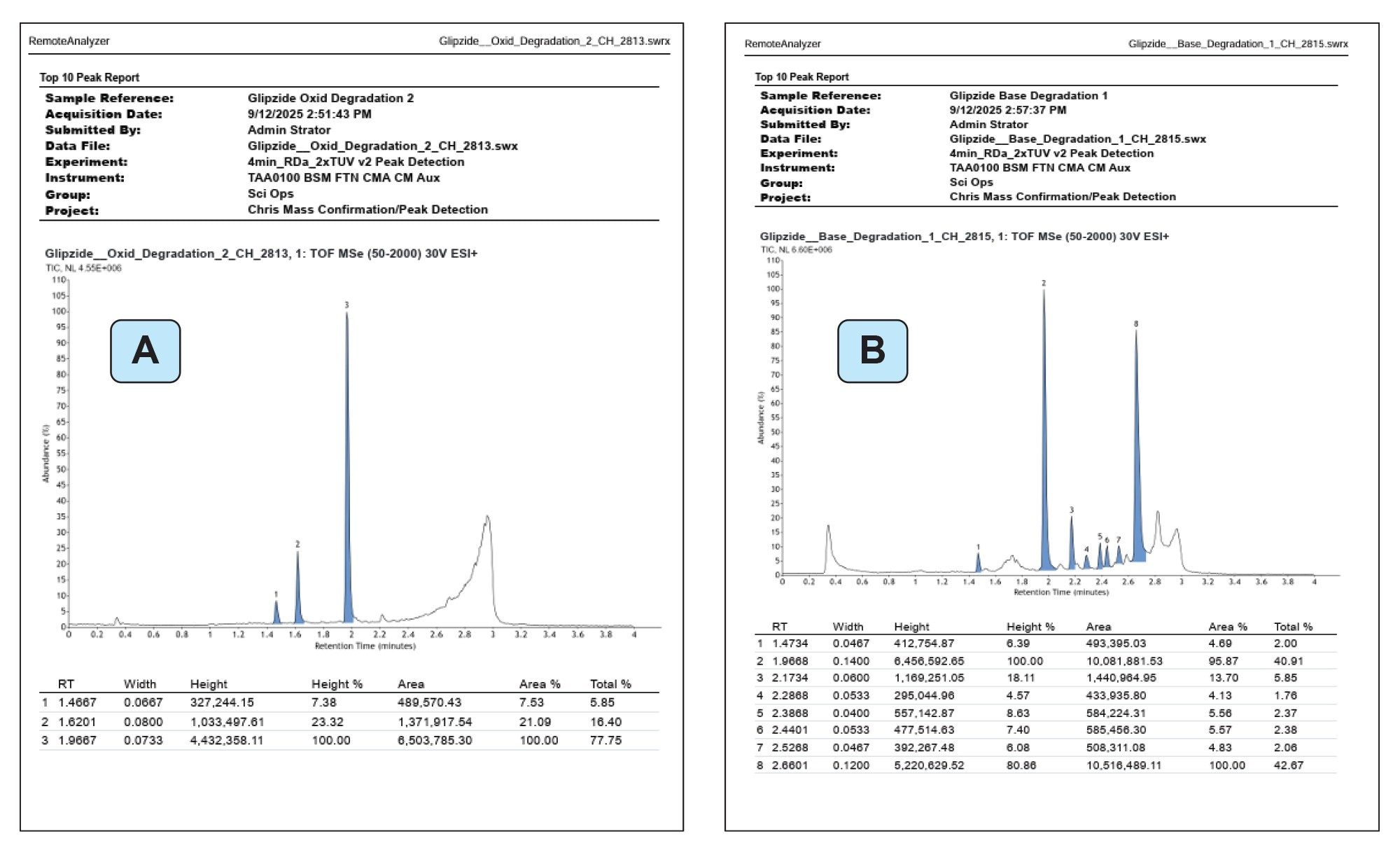
In this example, a total of three peaks were detected including the API glipizide. Similarly for the oxidation experiment, three peaks including the API were detected (Figure 2A) and for basic degradation eight peaks were detected (Figure 2B).
With the mass confirmation of the API successful, further interrogation of the data was required to attempt to identify the unknown peaks in the samples.
The acquired samples were easily downloaded from RemoteAnalyzer, packaged as an AnalyzerPro XD sequence so that these samples could be processed offline.
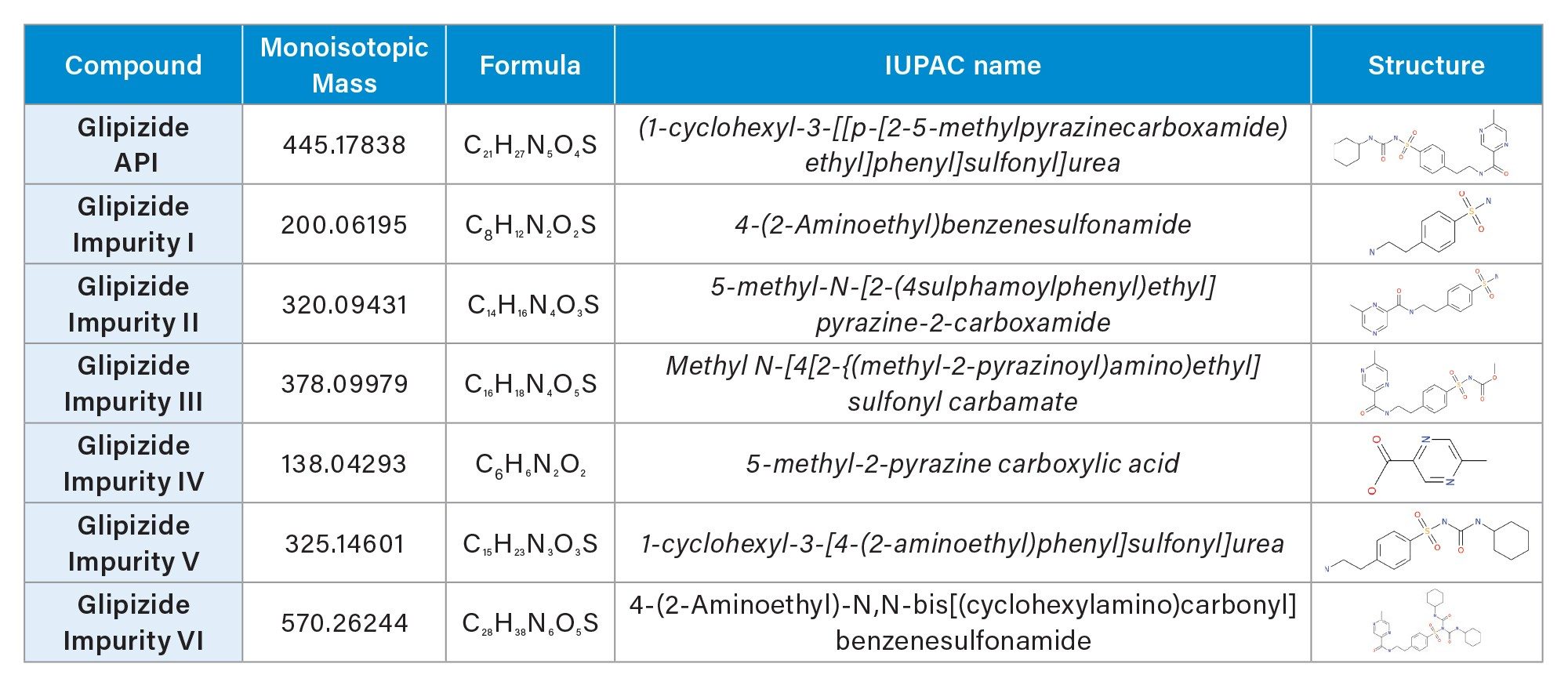
Once transferred, the data were interrogated using a processing method stipulating thresholds, adducts, ppm mass error etc. To assist with the identification of the unknown peaks, mol. Files for six known glipizide degradants (Table 1) as well as the API were incorporated into the processing method.
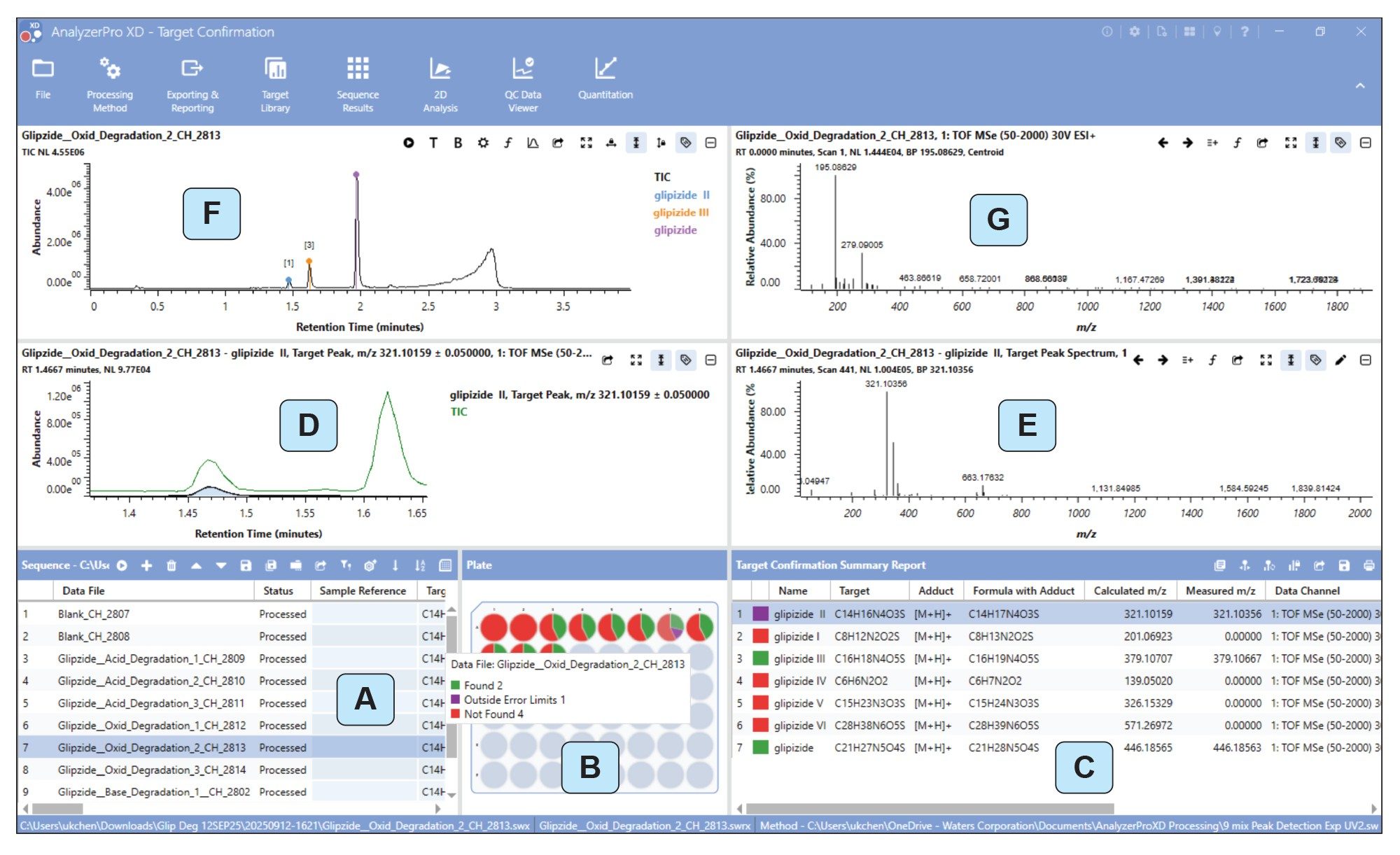
The AnalyzerPro XD data review page (Figure 3) presents multiple linked windows for scrolling through and interrogating data. (Figure 3A) lists the samples for processing and any targets that have been specified with the incorporation of molecular files or manual empirical formula. Selection of individual data files will be reflected in the data review windows. (Figure 3B) shows a graphical vial plate display with a color coded indicator corresponding to the Target Confirmation Summary Report i.e., ‘red’ signifies ‘not found’, ‘green’ signifies ‘found and within error limits’ and ‘purple’ signifies ‘found but outwith error limits’. Hovering the cursor over a vial/plate position gives a brief summary of target results.
(Figure 3C) gives a more comprehensive breakdown of the screened targets such as response, retention time, TIC alignment etc. (Figure 3D) displays the XIC for a selected identified target aligned with the trace specified in the processing method – here the TIC trace was used. The corresponding target spectrum is displayed in (Figure 3E). (Figure 3F) is a modifiable data review panel that allows individual/overlays of TIC/Base Peak/XIC’s and UV. (Figure 3F) displays the corresponding spectra of a selected scan in the TIC review window.
The data provided by AnalyzerPro XD are easily digestible, yet comprehensive, ensuring confidence in the assigned identifications.
All accurate mass measurements returned for the identified components across the samples were sub 5 ppm with the exception of impurity II in the second oxidation degradation which gave a ppm error of 6.14. This served as an indicator to the system administration that the instrument would benefit from recalibration. This was conducted by a simple click to recalibrate the instrument. Once calibrated, the samples were rerun, returning sub 5 ppm mass accuracy for all identified components for the oxidatively stressed samples (data not shown).
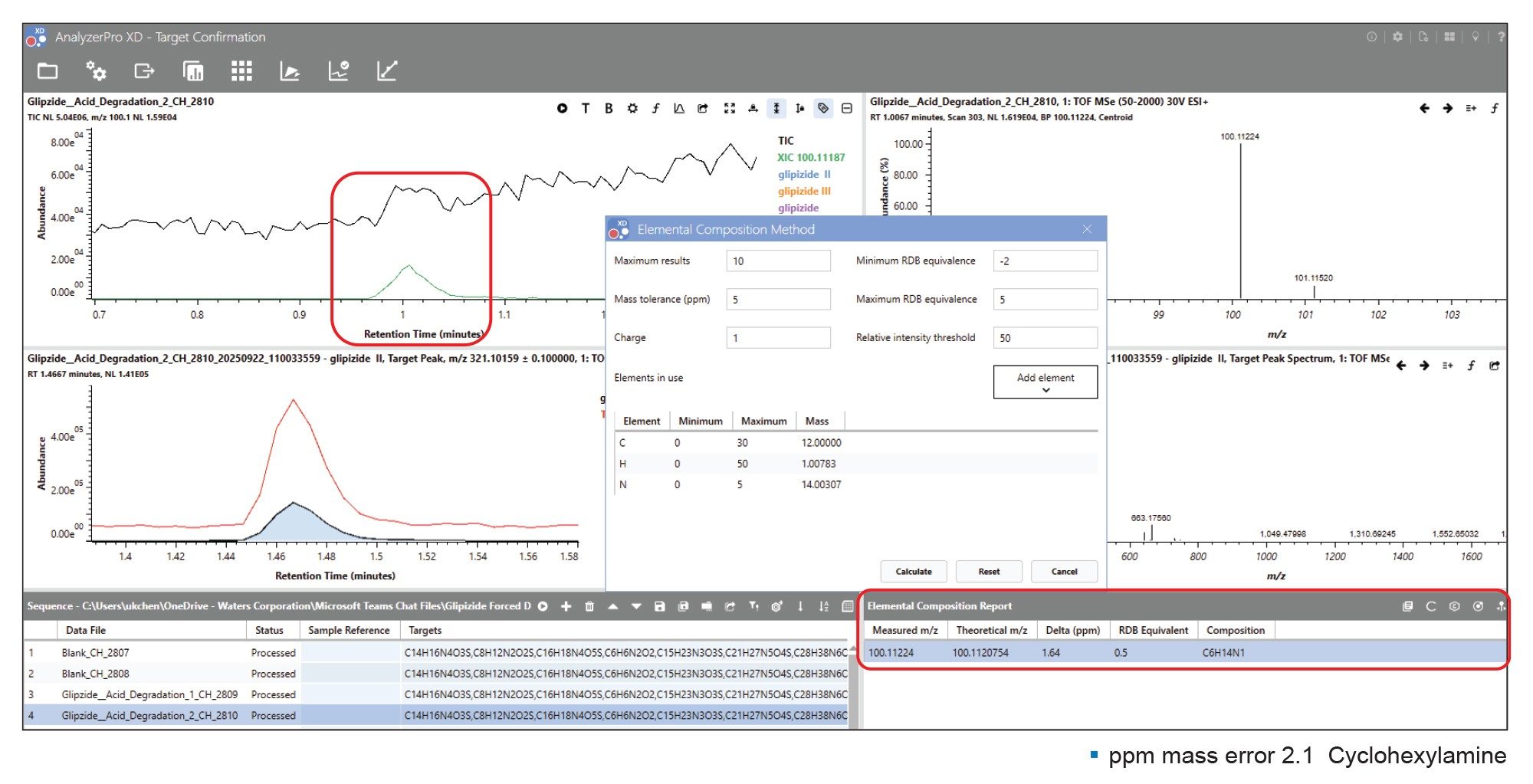
An unidentified peak was detected at a retention time of 1.00 minute, m/z 100.11224 in both the acidic and oxidatively stressed samples (Figure 4). Elemental composition was determined using the tool provided and was returned as C6H14N for the protonated compound. Compound assignment was performed by clicking the ‘PubChem’ button on the ‘Elemental Composition Report’ which directly linked to the website populating the search bar with the proposed formula. Searching the online library returned a hit for ‘cyclohexylamine’, a known glipizide impurity5 with a ppm mass error of 1.64.
Multiple peaks detected in the base degradation failed to return a compelling ‘hit’ either using molecular files or online library searching. These compounds may require identification using a specialist degradation pathway software e.g., Zenith™ (Lhasa Ltd., Leeds, UK) to adequately assign these peaks.
Conclusion
Drug discovery and development is an expensive and time-consuming process requiring streamlined operations to gather accurate data in as short a timeframe as possible. HRMS, with its ability to identify compounds more confidently through accurate mass, isotope patterns, better ion separation and detection, can help accelerate the discovery and development process.
With the ACQUITY RDa, access to accurate mass measurements is now available to non-HRMS experts. When combined with RemoteAnalyzer software, a flexible and intuitive sample submission interface minimizes sample submission time and provides useful in-depth sample information in an intuitive manner without the need for proficiency in complex MS informatics software.
Using this platform, a batch of chemically stressed glipizide samples were submitted in a single step. Targeted and untargeted workflows were used to screen the degraded sample profiles to mimic potential reaction mixtures seen in synthetic route design samples.
Each sample returned an EIC for the parent compound glipizide for all samples along with accurate mass measurements for the highest intensity peaks as defined in the processing method. Further processing in AnalyzerPro XD confirmed the presence of impurities II and III in the acid and oxidatively stressed samples. The presence of impurity V was confirmed in the base degraded sample.
References
- Termopoli V, Torrisi E, Famiglini G, Palma P, Zappia G, Cappiello A, Vandergrift GW, Zvekic M, Krogh ET, and Gill CG. Mass Spectrometry Based Approach for Organic Synthesis Monitoring Analytical Chemistry 2019 91 (18), 11916–11922 DOI: 10.1021/acs.analchem.9b02681.
- Fontana A, Iturrino L, Corens D, et al. Automated open-access liquid chromatography high resolution mass spectrometry to support drug discovery projects. J Pharmaceut Biomed Anal. 2020; 178:112908.
- Beccaria M, Cabooter D. Current developments in LC-MS for pharmaceutical analysis. Analyst. 2020;145(4):1129–1157.
- Henry C, Rainville P. Targeted and Non-Targeted Identification and Characterization of the Forced Degradation Products of Glipizide Using the ACQUITY RDa Detector and UNIFI Software Workflows. 720007510, February 2022.
- Albu M, Medvedovici A, Tache, F. (2010). HPLC/DAD Assay of Cyclohexanamine as Related Impurity in Glipizide Through Derivatization with o-Phtaldialdehyde. Analytical Letters - ANAL LETT. 43. 1162-1171. 10.1080/000327109035185.
720009089, November 2025