Analytical Scale 96-well Protein A Affinity Resin-Based Purification using Andrew+™ Automation Robot Supporting Upstream Bioprocessing
Yun W Alelyunasa, Julie Wushenskya, Mark Wronab, Rui Chena
a Waters IMMERSE™ Lab, Newark, DE, United States
b Waters Corporation, Milford, MA, United States
Published on September 05, 2025
Abstract
An automated Protein A affinity purification protocol for up to 96 culture media samples in a well plate format is described. The automation uses Cytiva™ antibody purification resin on Andrew+ Liquid Handling Robot. The purification consumes 120 µL HCCF to generate 100 µL of antibody in neutral pH buffer solution. For 96 samples, the purification protocol is comprised of a 55 minute liquid handling step and a 20 minute offline mixing during the Protein A binding step. The method was validated based on 96 samples each containing 173.5 µg of antibody. The purified amount as determined by LC-UV using Waters™ BioResolve™ Protein A Affinity Column was 167.7±14.2 µg, or recovery of 96.8±8.2%. Reproducibility of the 96-sample preparation was also excellent at ±8.5%. The Andrew+ automation protocol has been published in OneLab™ library and is available for download under the name “ProA-resin purification”.
Benefits
- Rapid and automated Protein A purification using Andrew+ Robot
- Automation protocol readily available for download in OneLab library
- Dynamic protocol to cover any number of samples from 1 to 96 in a well plate format
- Robust protocol with excellent recovery and reproducibility
- Excellent chromatogram characteristics of Waters BioResolve Protein A Affinity Column for the quantification of monoclonal antibodies
Introduction
Affinity purification of recombinant monoclonal antibody (mAb) refers to the use of a binding agent to extract an antibody from a complex matrix such as harvested cell culture fluid (HCCF). It is a reversable process where the binding occurs under physiological conditions and the antibody is released by a releasing reagent, typically under acidic conditions. The pH of the final solution is neutralized to maintain the antibody’s stability for long-term storage. In recombinant mAb production and analysis, Protein A (ProA) ligand, immobilized on a solid surface, is the most frequently used affinity agent.
In biotherapeutic process optimization, monitoring process- and product-related quality attributes are routinely performed in a typical BioPharm setting. The Ambr250® multi-parallel bioreactor system is a widely used platform for upstream bioprocess development. In a typical fed-batch experiment, a design of experiment (DoE) setup with sampling up to 14 days can generate a large number of samples. To enable time-course analysis of multiple product attributes efficiently and consistently, automated purification becomes a critical step of the overall workflow.
This application note describes automated purification using Andrew+ Robot and ProA ligand immobilized on resin for up to 96 HCCF samples in a 96-well plate format.1 It is a dynamic protocol, meaning when fewer than 96 samples are needed, the user simply enters the number of samples at the start of a run. The protocol will automatically adjust and prepare the specified number of samples. The recovery and repeatability of the automated purification was evaluated by LC-UV analysis using Waters BioResolve Protein A Affinity Column where ProA affinity ligand was immobilized on chromatographic media.
Experimental
HCCF Sample Generation
A commercially available NISTCHO cell line (RM8675 ext. NIST.gov) expressing mAb was cultured for 14 days to generate HCCF samples. Day 14 samples from multiple reactors were pulled, centrifuged, and filtered through 0.2 µm syringe filter. Samples were stored in a -80 °C freezer.
Sample Preparation for Recovery and Reproducibility
The HCCF sample was transferred into a 96-well plate and mAb was purified using Andrew+ Robot. The purified mAb and unpurified mAb in HCCF were subject to LC-UV analysis with NISTmAb (RM8671 ext. NIST.gov) as the calibration standard. All samples were injected in triplicate.
LC Method Conditions
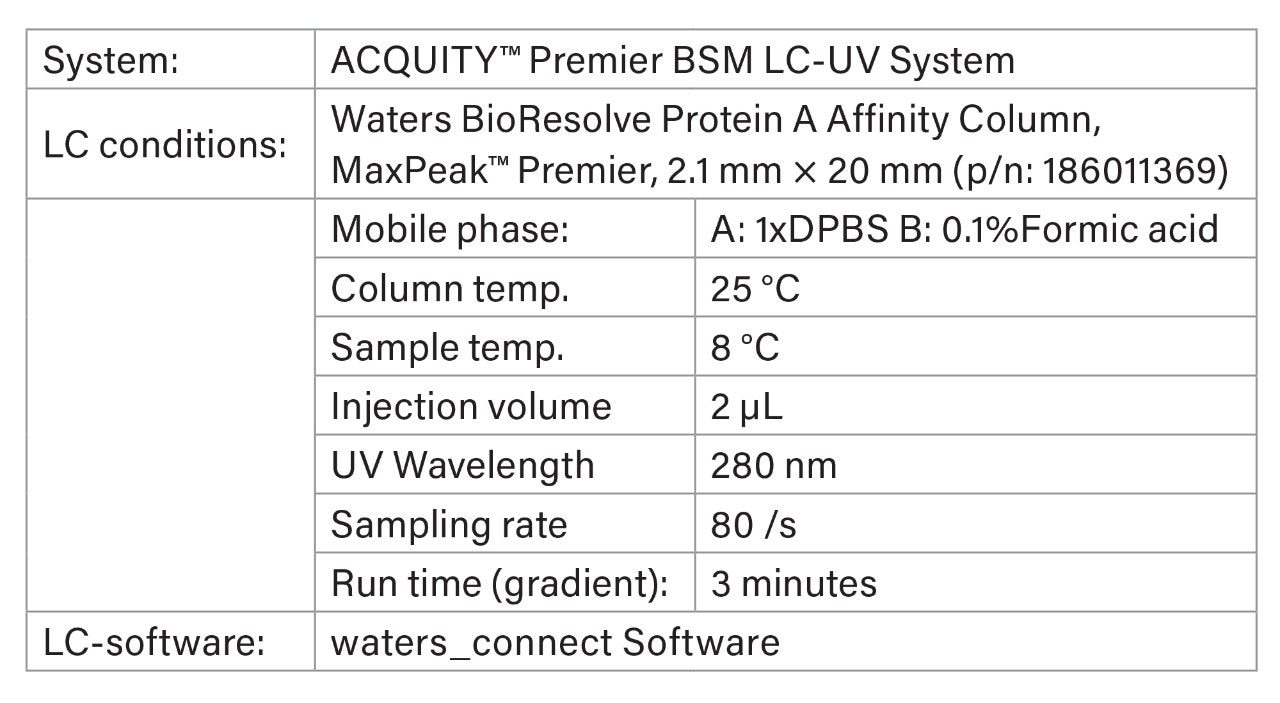
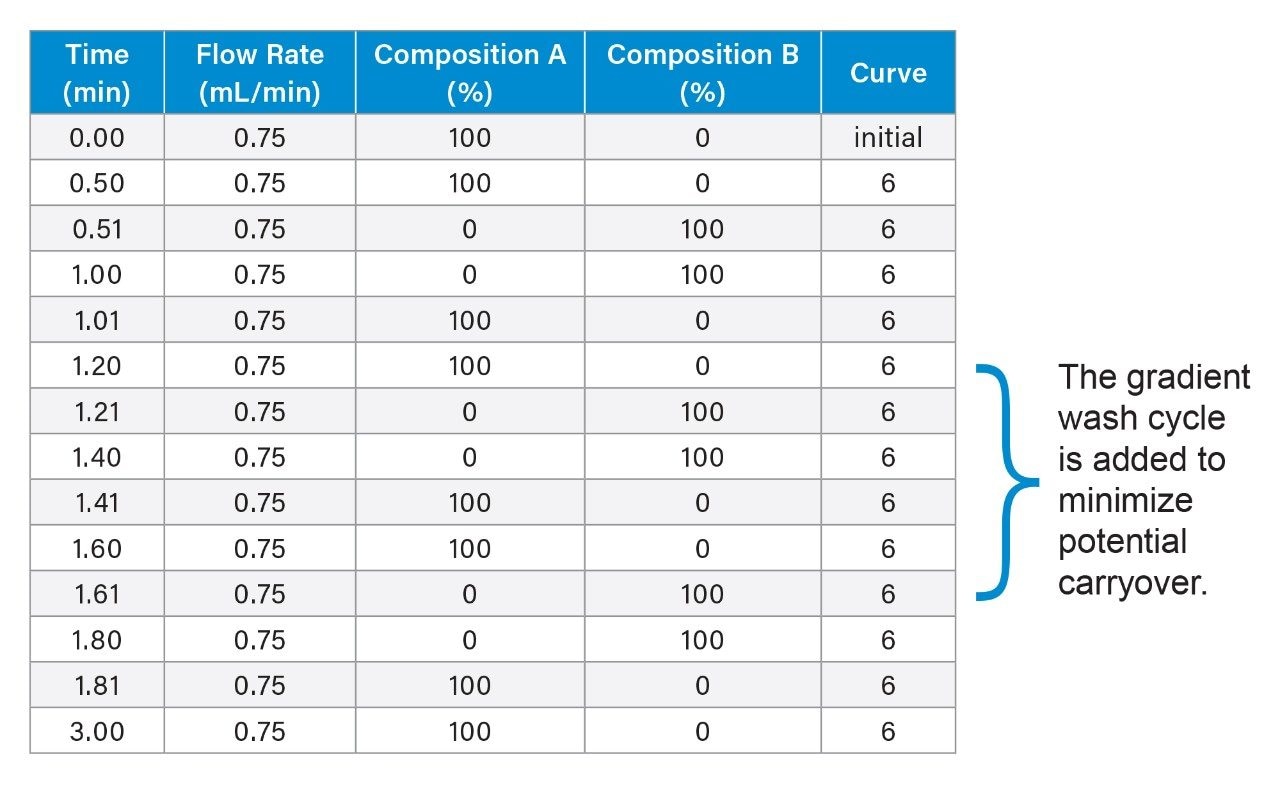
Gradient Table
Results and Discussion
I. Description of automated ProA affinity purification based on 96-well plate
A OneLab protocol for automated ProA affinity purification was developed for up to 96 samples in a 96-well plate format. For an Ambr250 System with 8 bioreactors and daily HCCF sampling over 14 days, one possible 96-sample layout is displayed in Figure 1.

The automated ProA affinity purification protocol used Cytiva MabSelect™ Antibody Purification Chromatography Resin. A general description for the OneLab protocol is summarized in Table 1 and the Andrew+ Robot deck layout is shown in Figure 2A. Briefly, Protein A resin was added to a filter plate and washed with water and phosphate buffer solution using an on-board Extraction+ vacuum filtration unit. After the bottom of the filter plate was covered with a film, HCCF solution was added. The top of the filter plate was then covered with a film and the plate was transferred to an offline Eppendorf® shaker and mixed for 20 minutes. After the mixing, both top and bottom films were removed, and the plate was returned to the Andrew+ Robot. The media solution was removed via vacuum filtration; the resulting resin with bound protein was washed with phosphate buffer and water. Finally, the mAb bound to ProA resin was released following three incubations with 100 mM glycine elution buffer at pH 3.0. The eluted mAb solution was neutralized with Tris buffer solution to pH 7.2 for protein stability and long-term storage.
The total recovered volume was 100 µL, approximately 20% enrichment of mAb concentration in media solution. The total time for the purification is 75 minutes for a plate of 96 samples. Excluding 20 minutes offline mixing using Eppendorf shaker, the total unattended liquid handling time was 55 minutes. The protocol is written as a dynamic protocol, allowing it to purify any number of samples from 1 to 96 samples in a well plate. Figure 2B shows the dialog box “Select the number of samples” that appears upon execution of the protocol where the number of samples to be prepared is entered. It is recommended to enter samples as multiples of eight to enable the use of 8-channel pipette for maximized speed. A detailed part list for Andrew+ Robot Dominoes and consumables are summarized in the Appendix.
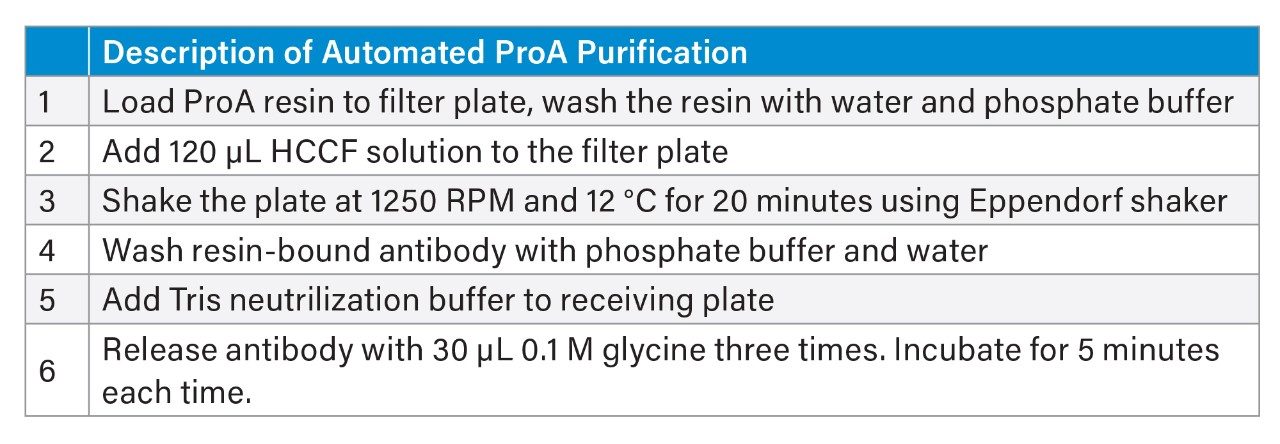
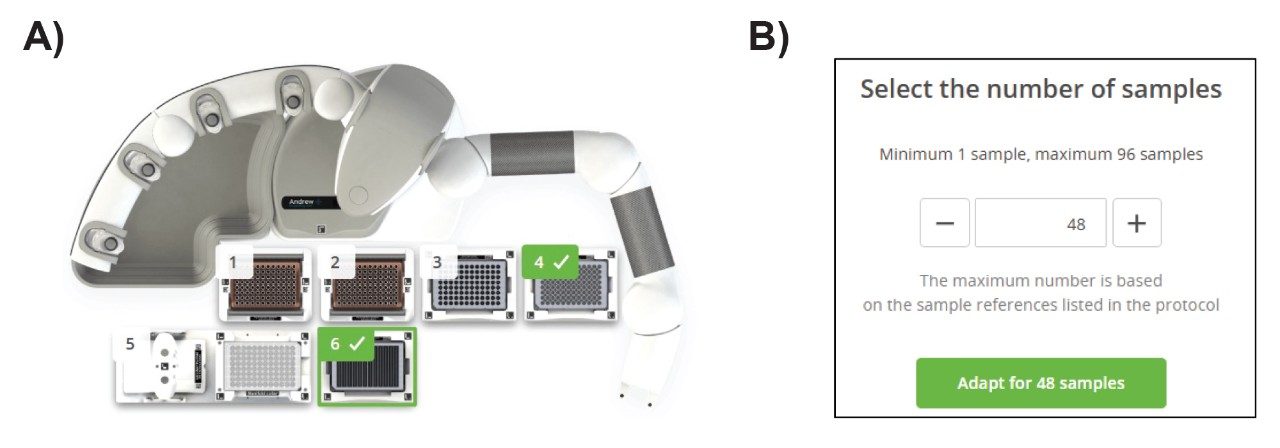
II. Sample recovery and reproducibility
The sample recovery was determined by LC-UV using Waters BioResolve Protein A Affinity Column. The samples used were 120 µL of pooled HCCF sample containing 1.44 mg/mL of mAb; the amount of antibody in the sample is 173 µg. For reference, the binding capacity for 60 µL of 25% resin slurry used in the present protocol is calculated to be 450 µg based on 30 mg/mL dynamic binding capacity from Cytiva’s product brochure.2
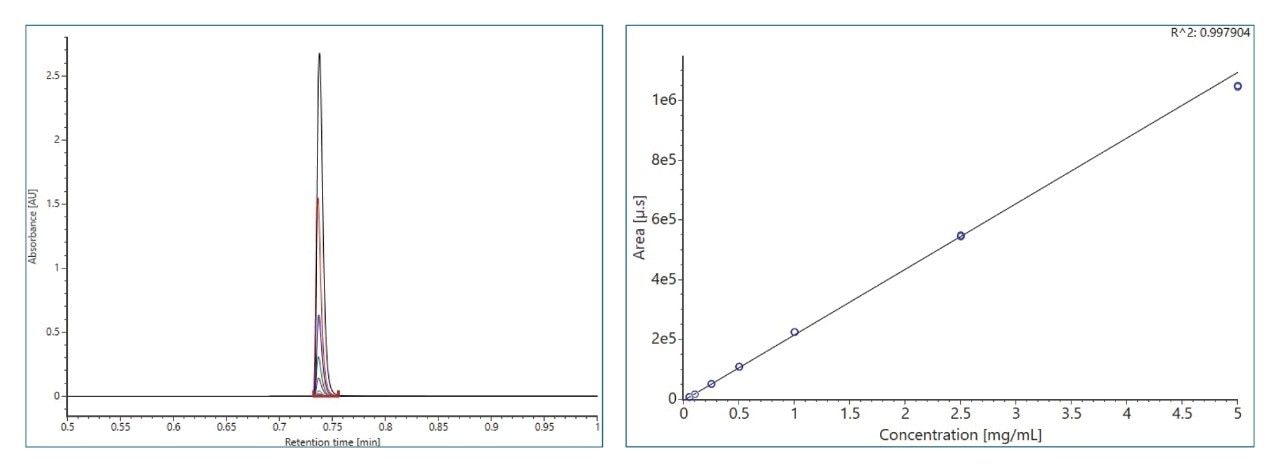
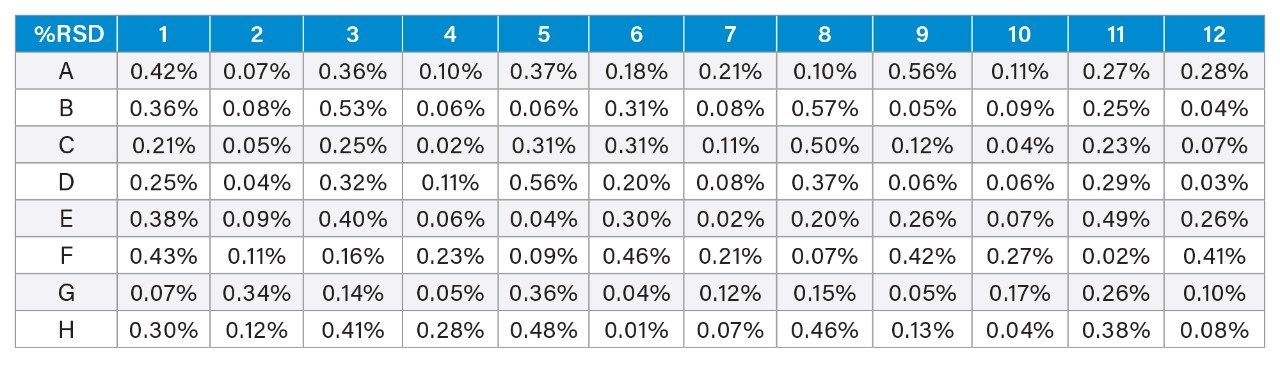
Figure 3 shows chromatograms and calibration curve of the NISTmAb standard solutions. At the prepared standard concentration range of 0.05–5 mg/mL, there is excellent linearity R2 = 0.998 and %deviation <10%. In general, the column produced excellent peak characteristics, 5x narrower peak width than other columns commonly used for titer determination.3 The sharp peak produced by the column resulted in excellent reproducibility for the triplicate injections used in sample analysis. Figure 4 shows %RSD of peak area from triplicate injections of the 96 samples analyzed. The observed %RSD for all samples is <1%.
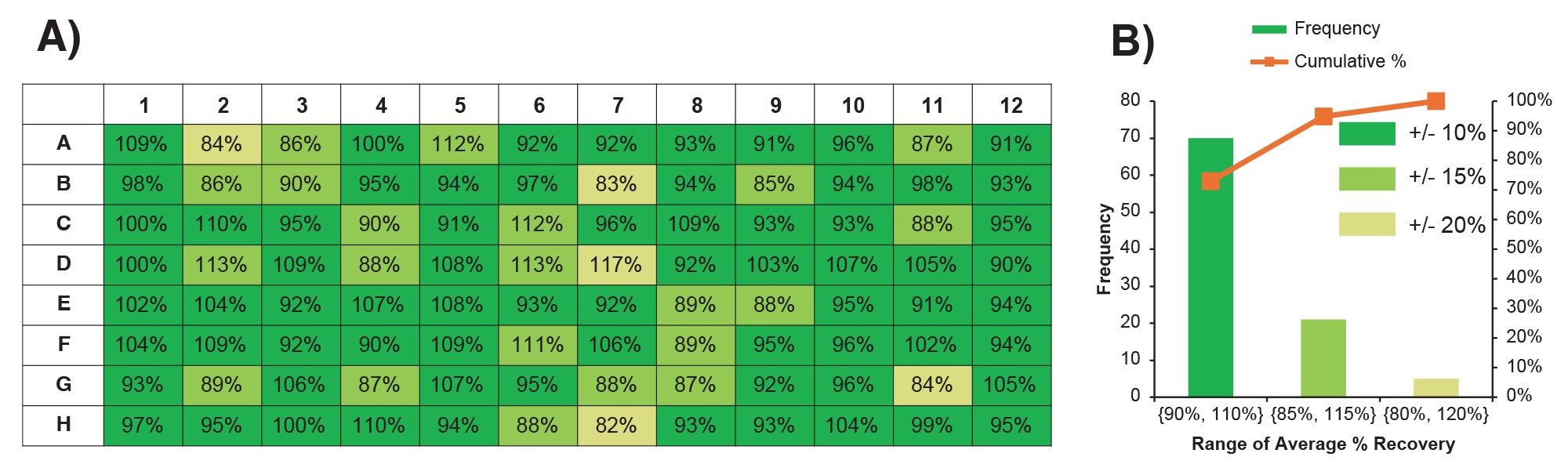
The mAb concentration was determined for the purified samples as well as the unpurified HCCF for the estimation of recovery of the automated purification. The determined concentration of mAb in purified sample is 1.68±0.14 µg/µL or the amount of 168±14 µg in 100 µL final volume. The reproducibility is excellent with 8.5% RSD for 96 samples. Compared to the amount of mAb in unpurified sample 173.4 µg, the overall recovery for 96 samples was calculated to be 96.8±8.2%. Figure 5A shows the average mAb recovery obtained per well in the 96-well plate format, and Figure 5B shows the histogram of the %recovery distribution. The data showed that the reproducibility of the %recovery for 70 out of 96 samples (73%) were within ±10%, 21 out of 96 samples (22%) were within ±15%, and only 5 out of 96 samples (5%) were within ±20%. Overall, the automated purification using Andrew+ Robot is robust and highly reproducible.
Conclusion
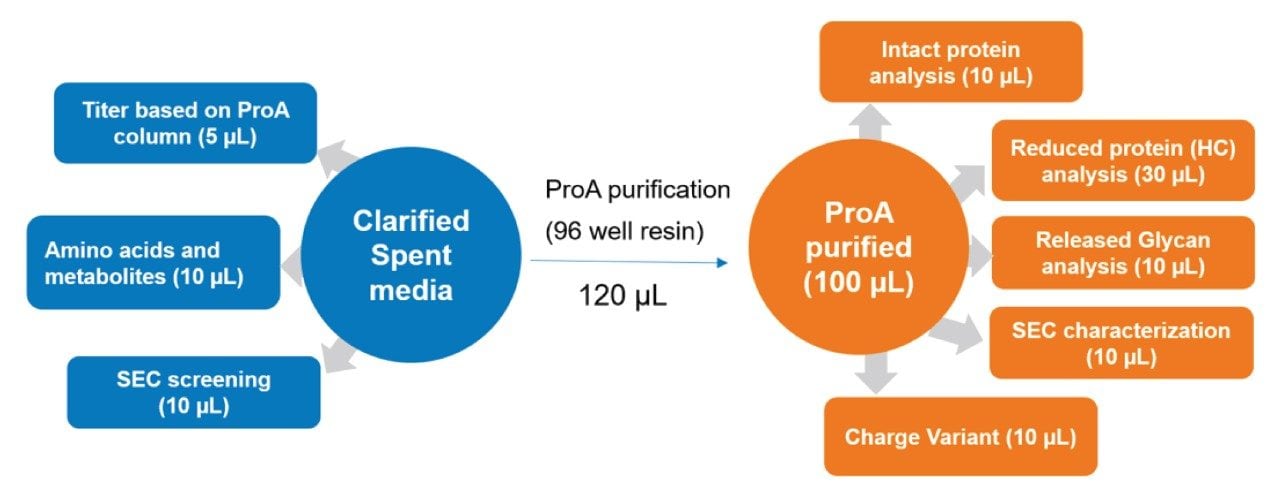
An automated ProA affinity purification protocol for 96 samples using Andrew+ Robot has been streamlined and uploaded to OneLab library. The protocol can be downloaded free of charge. The rapid purification produced a sufficient amount of antibody for multiple critical quality attribute (CQA) analyses, including intact protein, reduced protein analysis, released glycan profiling, size exclusion chromatography (SEC) and charge variant analysis, among others. An example of sample distribution from HCCF and ProA purified sample for various analysis is summarized in Figure 6.
Additional highlights of the automation protocol include:
- The total run time is 75 minutes. Excluding the offline plate shaking time of 20 minutes, the total unattended liquid handling time is 55 minutes.
- Minimal sample consumption (120 µL HCCF per sample).
- There is an excellent recovery of 96.8±8.2% for 96 samples preparation.
- The recovery is highly reproducible with RSD of 8.5% for 96 samples.
References
- Automated High-Throughput Analytical-Scale Monoclonal Antibody Purification Using Production-Scale Protein A Affinity Chromatography Resin, Stephan M. Koza Caitlin M. Hanna Albert H. W. Jiang Ying Qing Yu. Waters application note. 720007861. 2023.
- MabSelect™ antibody purification chromatography resin | Cytiva. Static binding capacity is not reported in the product literature.
- Lowering Quantitation Limits for mAb Titer Measurements Using Small Volume 3.5 µm Particle-Size Protein-A Affinity Columns, Stephan M. Koza, Steve Shiner, Matthew A. Lauber. Waters application note. 720008775. 2025.
Appendix
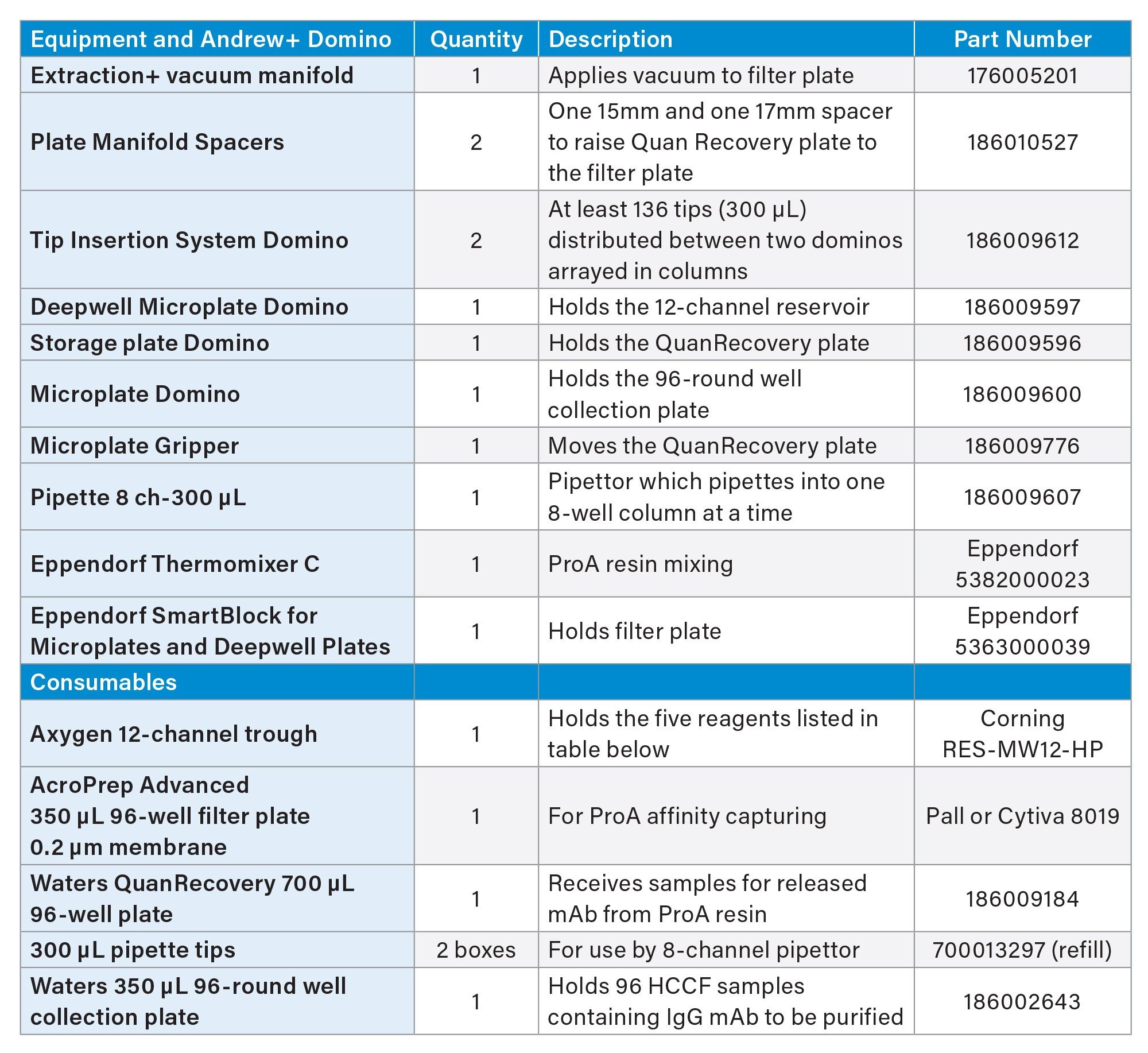
Table 2 is a summary list of Dominos and consumables used in ProA affinity purification using Andrew+ Robot. Table 3 is a summary of reagents used and their placement in reagent trough (Figure 7).
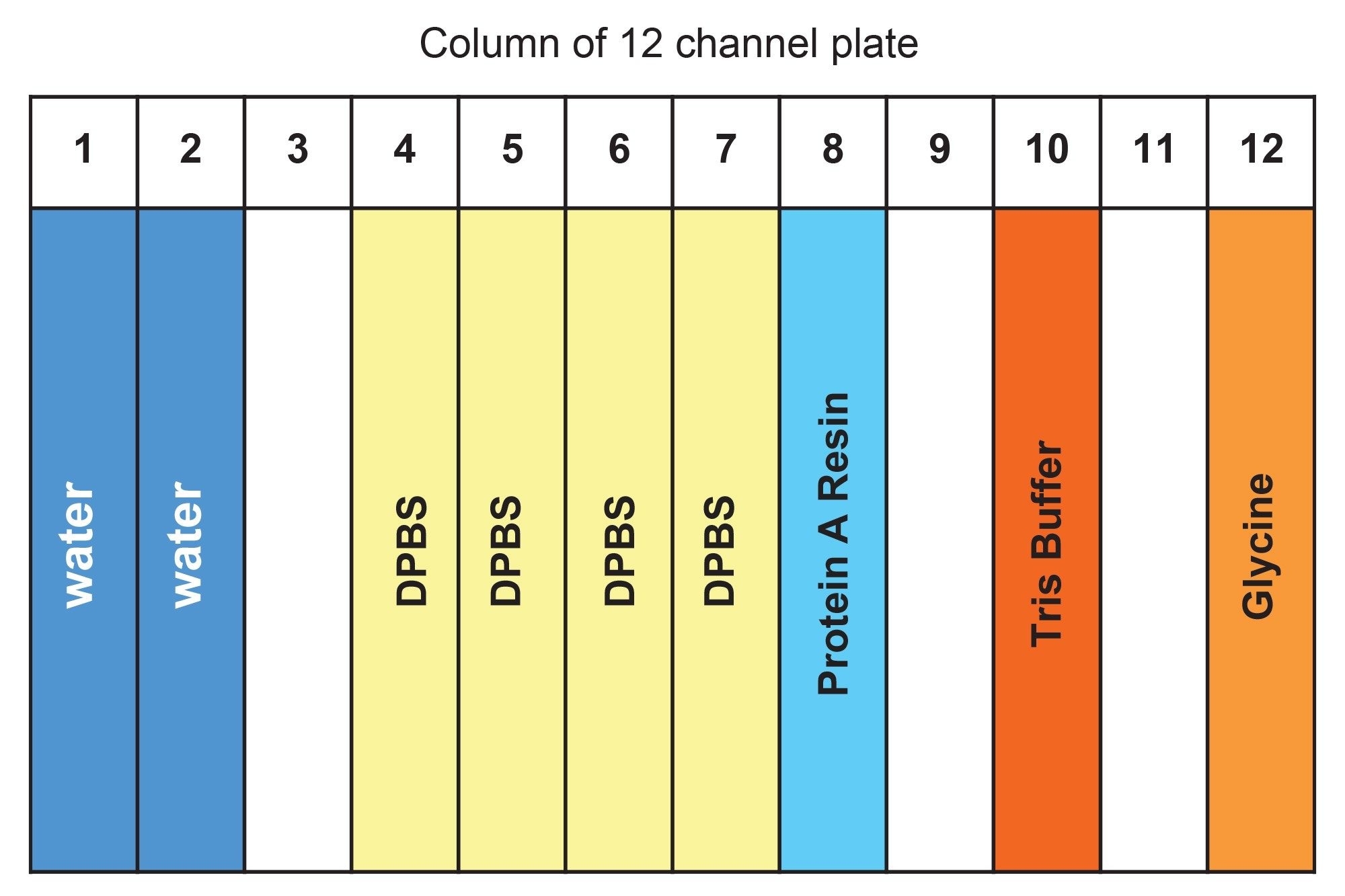
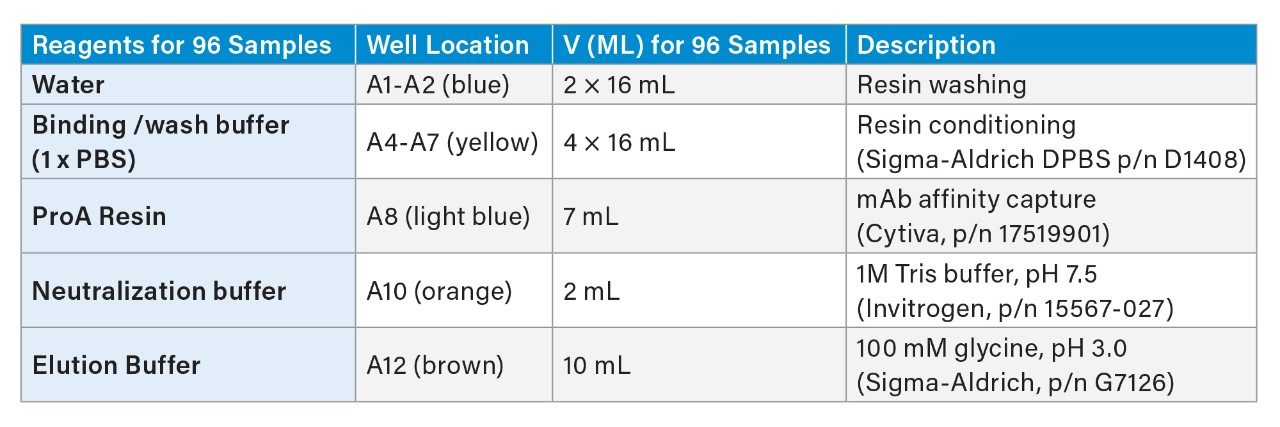
Instruction for reagent preparation
- Binding/wash buffer: 1xDPBS, perform 1:10 dilution of 10xPBS (Ex. Sigma-Aldrich) using H2O
- ProA resin: The resin comes as 50% resin in 20% ethanol; following Cytiva’s instruction, centrifuge at 1000 g for 3 minutes, then replace supernatant with 400 mM NaCl in 20% ethanol to 50% resin level. Further dilute with 1xDPBS to 25% resin as the working solution. Thoroughly mix prior to transfer to reagent trough. After each use, mark the liquid level on the tube. In next use, if liquid level is reduced due to evaporation, fill to mark with 20% ethanol, 0.4 M NaCl solution.
- Elution buffer: 100 mM glycine, pH adjusted to 3.0 using 5 N HCl.
- Neutralization buffer: 1 M Tris-HCl, pH 7.5.
Featured Products
720009002, August 2025