Structural Characterization of Glycated Monoclonal Antibodies Using Collision Induced Unfolding on the SELECT SERIES™ Cyclic™ IMS Mass Spectrometer
Abstract
Mass spectrometry-based methods are the gold standard for the characterization of novel biopharmaceutical products. Most frequently in conjunction with chromatographic separations, the methods primarily focus on the determination of amino acid sequence, post-translation modification profiles, and degradation products. In contrast, the elucidation of the factors that contribute to the folding and stability of native biomolecules is traditionally performed using calorimetric techniques such as differential scanning calorimetry. The understanding of thermodynamic stability and folding mechanisms is highly important in biotherapeutic developability assessments. In this application note we demonstrate the application of native ion mobility-mass spectrometry (IM-MS) and collision-induced unfolding (CIU), a gas phase calorimetric technique, for protein unfolding and stability measurements of glycated monoclonal antibodies. Using the NISTmAb reference material, IM-MS-based collision-induced unfolding measurements and the application of CIUSuite3 software, we observe the effect of glycation on mAb structure by probing the gas phase unfolding pathway of the protein. The results demonstrate the potential of CIU as a high throughput technique to evaluate protein folding and stability in a solvent free environment while assessing the developability of a biotherapeutic molecule.
Benefits
- Measure protein unfolding pathways in minutes with collision induced unfolding (CIU) and Cyclic ion mobility
- Fully integrate the CIU workflow with CIUSuite3 software to perform higher order structural stability studies
- Increase confidence in results by reducing interference with pre-IMS quadrupole selection
Introduction
Mass spectrometry methods are central to the characterization of the primary structure of biopharmaceutical proteins. Intact mass and peptide mapping workflows give detailed information on critical quality attributes (CQAs) that report on amino acid sequence, glycosylation profiles, and degradation products such as truncations, oxidations and glycations. Such CQAs can influence the safety and efficacy of biotherapeutics and so must be monitored and controlled, but increasingly there are efforts to understand the effects of these CQAs through measurements of higher order protein structure.
Measurements of higher order structure can be probed in detail with MS-based methods such as hydrogen/deuterium exchange MS, chemical crosslinking and native ion mobility mass spectrometry (IM-MS). The latter is growing in popularity in the biopharmaceutical industry due to its rapid nature and ability to report on protein:protein and protein:ligand interactions as well as protein folding. The ability of IM-MS to probe protein conformations is well studied, but often structural changes are too subtle for outright resolution of different conformational families for characterization. Collision-induced unfolding is a branch of native IM-MS in which gas phase protein ions are unfolded by imparting energy through acceleration of ions into an inert bath gas (Figure 1A). The technique has been shown to report on ligand-induced stabilization(1,2), changes in domain structure(3,4), disulfide bonding pattern(5) and glycosylation profile(6).1–6 Unfolding transitions can be monitored by mobility separation of emerging conformers as the energy of the collisions is increased (Figure 1B). By monitoring the arrival time distribution of the unfolding ions as a function of collision voltage, a ‘CIU fingerprint’ can be constructed to describe the entire unfolding pathway. By comparing CIU fingerprints of different conditions, e.g. stressed versus unstressed or apo versus holo, subtle effects on protein structure and stability can be revealed.
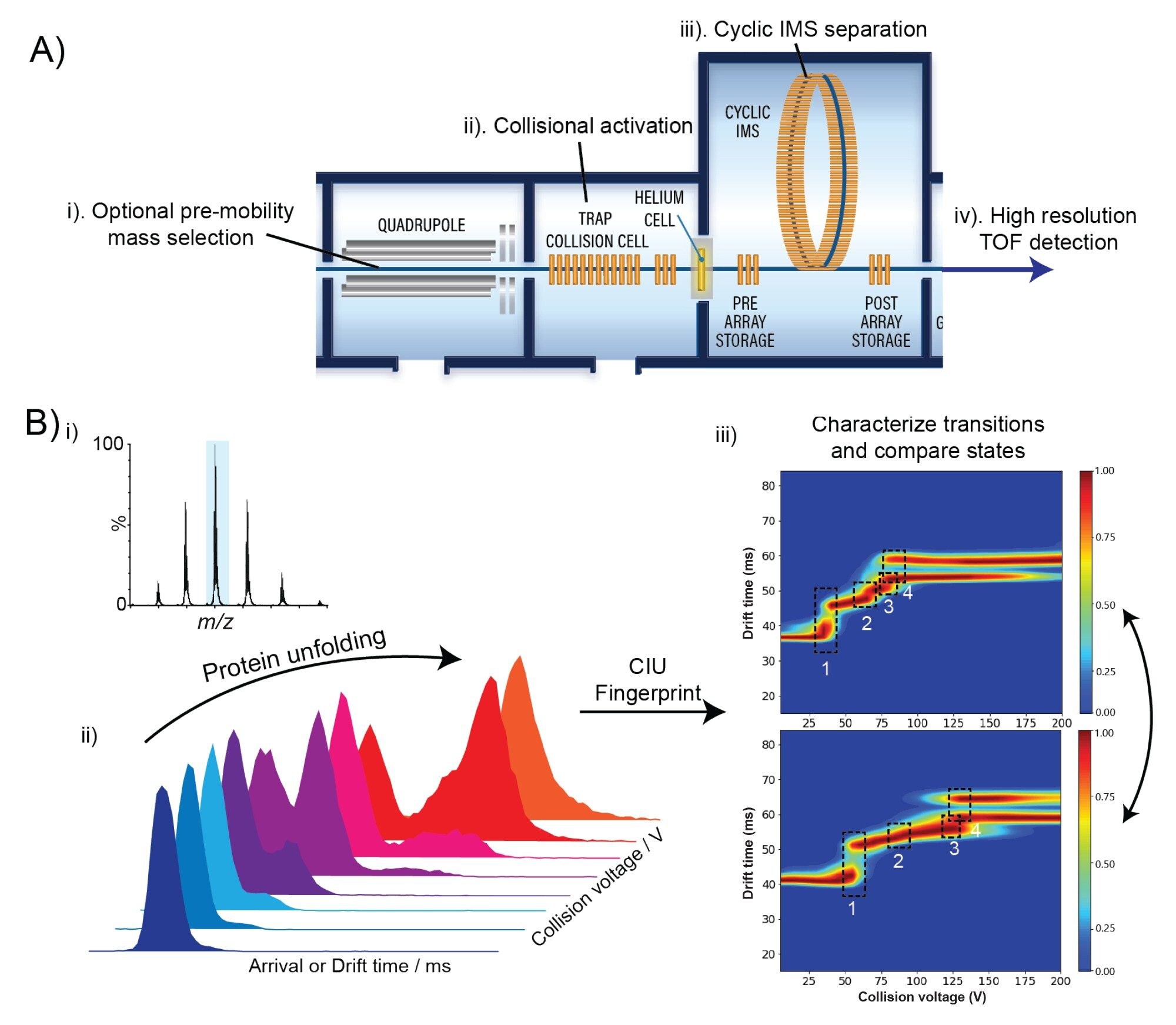
Experimental
Sample Description
NISTmAb RM8671 was purchased from NIST. The protein was received at 10 mg/mL in 12.5 mM L-histidine buffer and then buffer exchanged into 200 mM ammonium acetate using 0.5 mL Zeba Spin benchtop centrifugal desalting column (ThermoFisher Scientific). The protein was diluted to 0.1 mg/mL using the ammonium acetate. Glycation reactions were performed with NISTmAb at 5 mg/mL in a solution with 1 M glucose at 30 °C for 30 minutes, 7 days and 16 days. Upon completion of the incubation the solution was buffer exchanged into water and then frozen. The control (30 minute timepoint) was immediately buffer exchanged into water and then frozen at -70 °C. All time points were buffer exchanged into ammonium acetate as previously described and diluted to 0.1 mg/mL prior to analysis on the mass spectrometer.
Method Conditions
NISTmAb samples were introduced into the mass spectrometer using long, thin-walled borosilicate glass nanocapillaries (p/n: M956232AD1). For native folded measurements the cone voltage and collision voltages were minimized.
MS Conditions
|
MS system: |
SELECT SERIES Cyclic IMS Mass Spectrometer |
|
Ionization source: |
Nanolockspray with static needle |
|
Ionization mode: |
Positive |
|
Acquisition range: |
50–8,000 m/z |
|
Capillary voltage: |
1 kV |
|
Trap collision voltage (no unfolding): |
5 V |
|
Trap collision voltage step for CIU: |
5 V |
|
Maximum trap voltage used: |
200 V |
|
Cone voltage: |
50 V |
|
Cyclic static wave height: |
25 V |
|
Pushes per bin: |
2 |
|
Array offset (inject step): |
60 V |
|
Array wave height (inject): |
4 V |
|
Array offset (separate): |
70 V |
|
Racetrack bias: |
70 V |
|
Array wave height (eject and acquire): |
25 V |
Data Management
|
MS software: |
Waters™ Embedded Analyzer Platform for Cyclic IMS v12.2 |
|
Informatics: |
Masslynx™ v4.2, Driftscope v3.0, CIUSuite3 (github.com/RuotoloLab/CIUSuite3) |
Results and Discussion
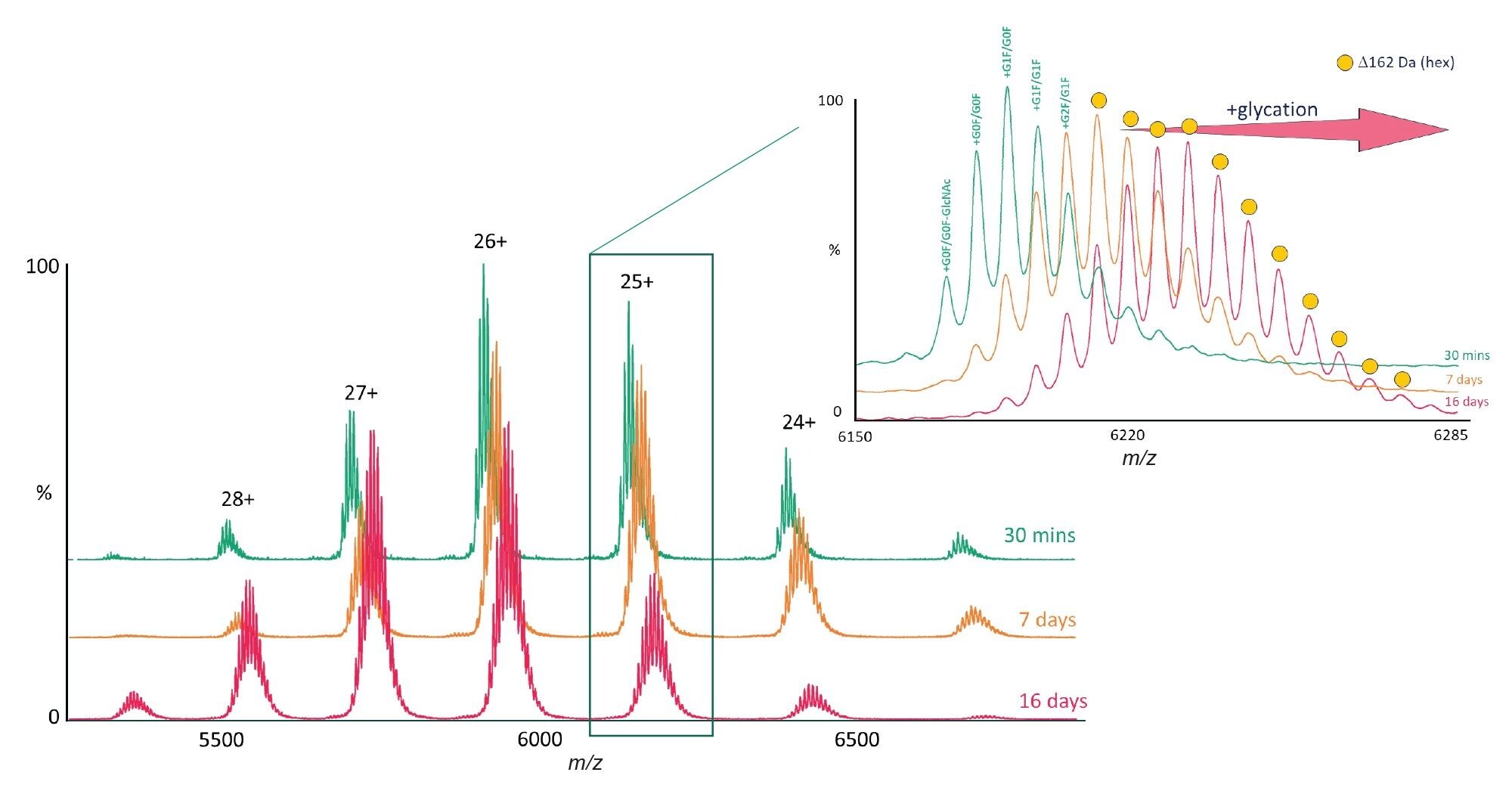
To investigate the effects of glycation on the unfolding pathway of mAbs we incubated the NISTmAb RM8671 protein with glucose for different time periods, namely 30 minutes, 7 days, and 16 days. Firstly, to confirm the degree of glycation we performed a native MS experiment on each of the time points (Figure 2). The mass spectrum after 30 minutes exhibited a small but detectable increase in the amount of hexose (+162 Da) over the non-incubated NISTmAb mAb. The spectrum of the mAb incubated for 7 days exhibited significant glycation with up to 8 hexose-modified proteoforms. The spectrum of the mAb incubated for 16 days exhibited additional modifications with greater than 11 hexose-modified proteoforms. Having confirmed the formation of glycation, CIU experiments were performed to investigate any changes in protein unfolding.
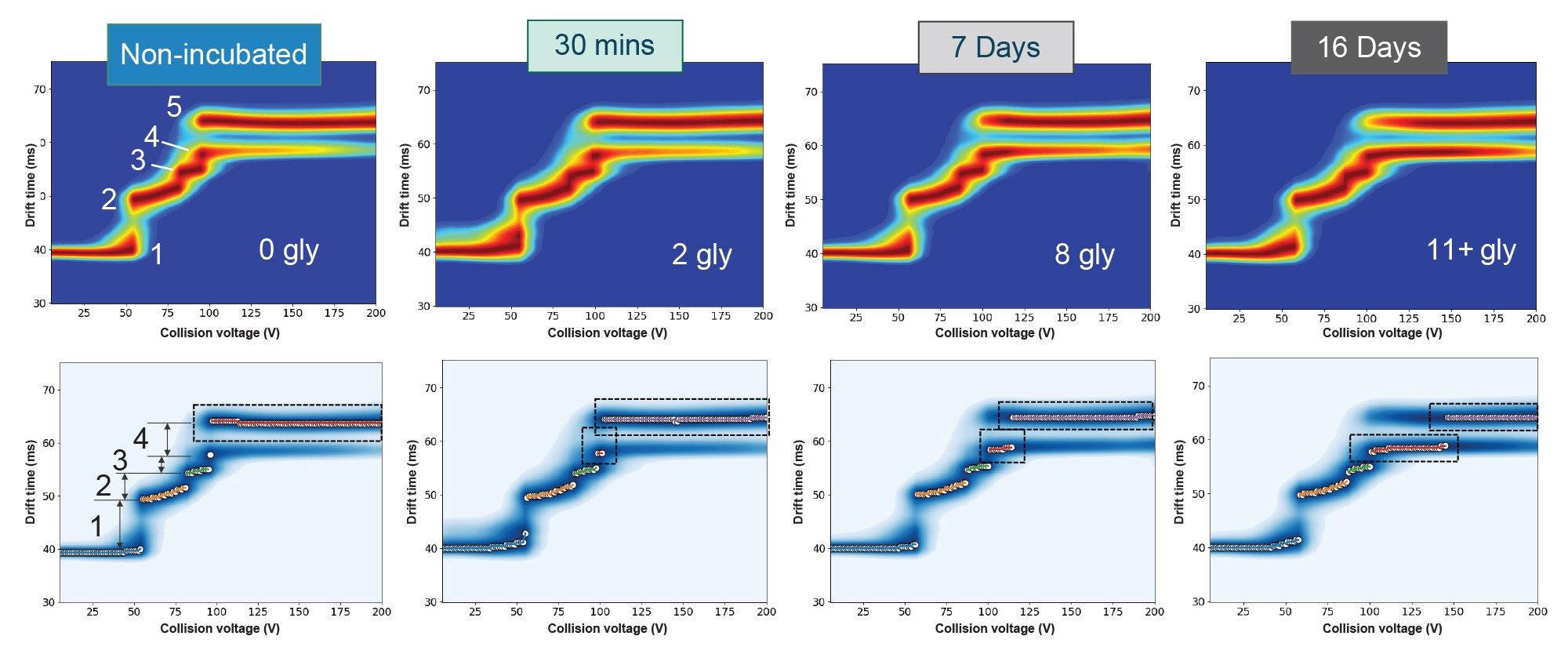
CIU experiments were performed on the non-incubated protein, 30 minutes-, 7 days-, and 16 days-incubated samples. For simplicity we show only the data for the 25+ charge state. To visualize the data, we chose to use CIUSuite3, the latest version of a popular software tool developed by the group of Professor Brandon Ruotolo at the University of Michigan, USA(7), that allows native reading of data from the SELECT SERIES Cyclic IMS instrument. Figure 3 (top row) shows the CIU fingerprints, generated in CIUSuite3, for all the states studied. The non-incubated protein shows five major states and four transitions. The positions of the transitions along the voltage axis reveal differences in stability (Figure 3 bottom row). As the level of glycation increases, the fourth unfolding intermediate persists for an increased range in collision voltage, resulting in the transition to the final unfolding intermediate occurring at a higher collision voltage. This observation demonstrates a stabilizing effect due to glycation based on the final transition requiring higher energy to fully unfold.
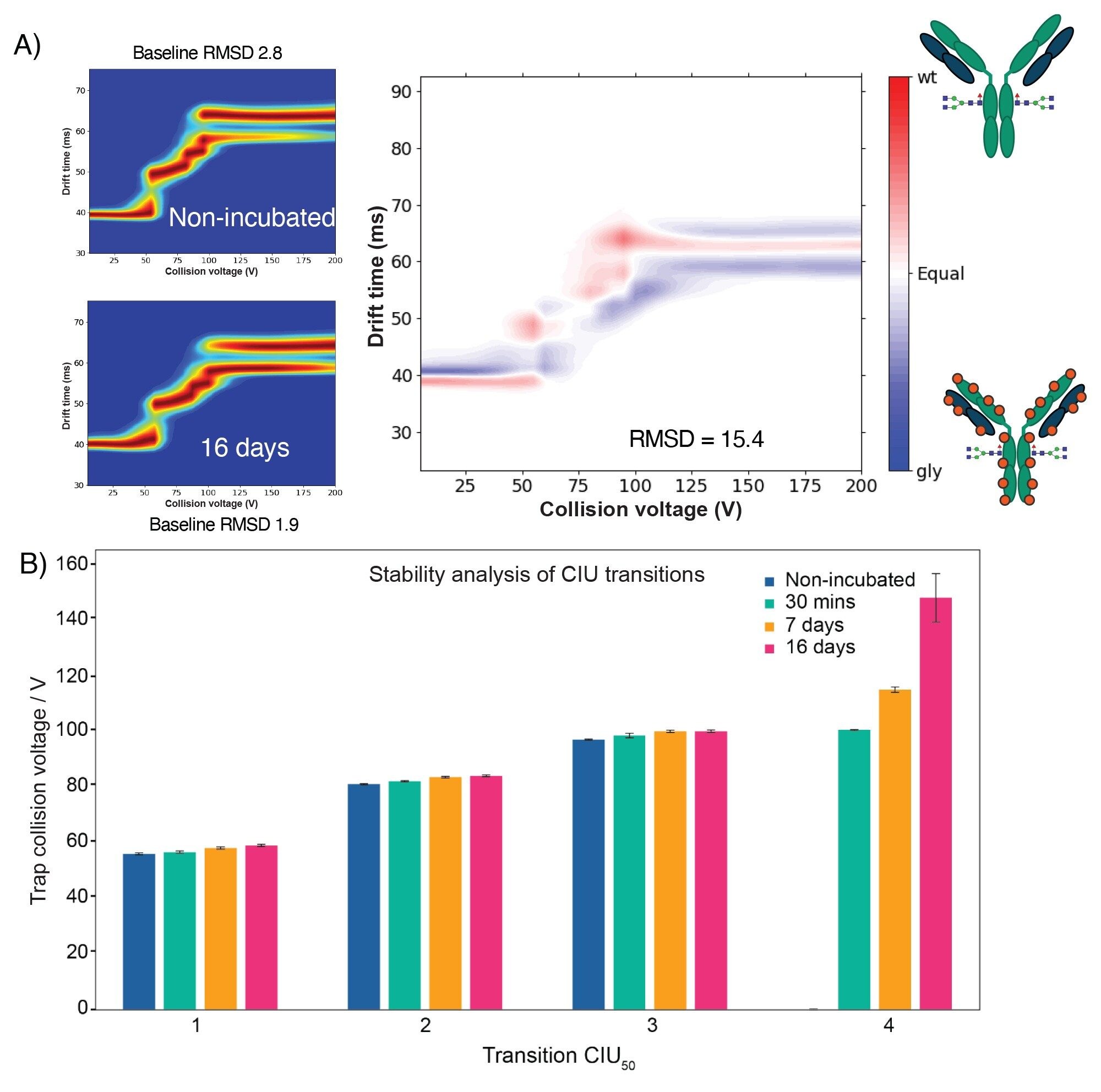
The tools within CIUSuite3 enable analysis of the CIU fingerprints by numerical means, removing user bias. Firstly, two conditions can be compared to quantify the difference between them. In this way we can assess the overall effect on the unfolding profile between the two conditions (Figure 4A). To this end, we compared the CIU fingerprints of non-incubated NISTmAb with the 16 days-incubated sample. A difference plot (Figure 4A right) shows the areas where the CIU fingerprints are more intense in either condition. Calculation of a root-mean-square deviation of the difference plot gives a numerical value by which to quantify the differences between the two fingerprints, in this case a value of 15.4 was obtained. Compared to the intra-condition RMSD of 2.8 between replicates, this is highly significant. Secondly, CIUSuite3 enables tracking of each unfolding transition independently, giving ‘CIU50’ values for each, which again can be compared between conditions (Figure 4B). A CIU50 value is measured as the collision voltage of the inflection point from where one feature transitions to the next, this value can provide insights on protein stability. Indeed, the graph in Figure 4B shows that transitions 1 to 3 are affected only slightly by the increase in glycation as CIU50 values are similar between each condition. Strikingly, however, transition 4 is drastically affected by the increase in glycation, with CIU50 values of 102, 115, and 148 V for 30 minutes, 7 days, and 16 days, respectively. These data support the observation of stabilization of transition 4 with increasing glycation.
Conclusion
In this application note we have shown the power of the SELECT SERIES Cyclic IMS mass spectrometer for the detection of structural changes in mAbs. Using the simple and rapid technique of CIU, the high-quality data obtained from the instrument enabled the characterization of the gas phase unfolding profile for the standard protein NISTmAb. The utility of the CIU approach was demonstrated when comparing the CIU fingerprints of NISTmAb to glycated NISTmAb showing a clear stabilization of its final unfolding transition with increasing amounts of glycation. The CIU workflow was enhanced with the CIUSuite3 software package, which enables native reading of data from the SELECT SERIES Cyclic IMS instrument and quantitative analysis of the CIU fingerprint. This powerful workflow provides a way for the structural integrity of biopharmaceutical products to be assessed rapidly during development, with the potential to save time and triage constructs and formulations more efficiently.
References
- Beveridge R, Migas LG, Payne KAP, Scrutton NS, Leys D, Barran PE. Mass Spectrometry Locates Local and Allosteric Conformational Changes That Occur on Cofactor Binding. Nat Commun. 2016. Nov;7(1):12163.
- Zhao B, Zhuang X, Bian X, Liu S, Liu Z, Song F. Stabilities of Superoxide Dismutase and Metal-Free Superoxide Dismutase Studied by Electrospray Ionization Ion Mobility Mass Spectrometry. Rapid Communications in Mass Spectrometry. 2019. 33(9):894–6.
- Zhong Y, Han L, Ruotolo BT. Collisional and Coulombic Unfolding of Gas-Phase Proteins: High Correlation to Their Domain Structures in Solution. Angewandte Chemie International Edition. 2014. Aug 25;53(35):9209–12.
- Watanabe Y, Vasiljevic S, Allen JD, Seabright GE, Duyvesteyn HME, Doores KJ, et al. Signature of Antibody Domain Exchange by Native Mass Spectrometry and Collision-Induced Unfolding. Anal Chem. 2018. Jun 19;90(12):7325–31.
- Tian Y, Han L, Buckner AC, Ruotolo BT. Collision Induced Unfolding of Intact Antibodies: Rapid Characterization of Disulfide Bonding Patterns, Glycosylation, and Structures. Anal Chem. 2015. Nov 17;87(22):11509–15.
- Tian Y, Ruotolo BT. Collision Induced Unfolding Detects Subtle Differences in Intact Antibody Glycoforms and Associated Fragments. International Journal of Mass Spectrometry. 2018. Feb 1;425:1–9.
- Jeon CK, Rojas Ramirez C, Makey DM, Kurulugama RT, Ruotolo BT. CIUSuite 3: Next-Generation CCS Calibration and Automated Data Analysis Tools for Gas-Phase Protein Unfolding Data. J Am Soc Mass Spectrom. 2024. Aug 7;35(8):1865–74.
Acknowledgement
We thank the authors Dr Kristine Parson, Mrs Margo Wilson, Dr Greg Adams and Dr Jason Barker from FUJIFILM Diosynth Biotechnologies for their valuable contributions to this application note.
Featured Products
720008668, December 2024