This is an Application Brief and does not contain a detailed Experimental section.
This application brief demonstrates the flexibility of desorption electrospray ionization (DESI) imaging, showing that the same tissue can repeatedly be imaged at the molecular level in positive and negative ion mode, to maximize information about molecular distribution in tissue.
To demonstrate the flexibility of desorption electrospray ionization (DESI) imaging, showing that the same tissue can repeatedly be imaged at the molecular level in positive and negative ion mode, to maximize information about molecular distribution in tissue.
DESI imaging, a surface analysis technique incorporating an electrospray probe, can be utilized as an imaging technique by rastering a surface under an ionizing solvent sprayer using a high precision X,Y stage. As the droplets impact upon the surface, chemical constituents are desorbed and carried towards the atmospheric inlet of the mass spectrometer. Ionization of the desorbed molecules occurs via the charge imparted onto the droplets.
In contrast to other ionization techniques often used for tissue imaging (i.e., MALDI), no special sample preparation, such as coating of the tissue section with a specialized solvent/ionizable matrix mixture, is required. In this study, gas and solvent flow rates as well as ionization voltages are optimized to allow DESI imaging experiments that preserve the tissue sample being analyzed. These less destructive DESI ionization conditions provide an opportunity for a single tissue section to be analyzed multiple times with the same or different experimental conditions or techniques.
Here we demonstrate that by optimizing DESI conditions, a wealth of molecular information can be accessed from a single tissue section, without the need to substantially alter the analysis conditions.
Mass spec imaging of tissue sections was accomplished by using a SYNAPT G2-Si HDMS Mass Spectrometer equipped with a 2D-DESI source. Data was generated and analyzed using HDI Software v1.3.
Fresh frozen tissues of porcine and human liver were sectioned on a cryo-microtome to 15 µm thickness and thaw-mounted onto conventional glass slides. When required, the samples were stored at -80 °C. Immediately prior to analysis, the samples were brought to room temperature and placed directly onto the stage of the DESI source. No further sample preparation was required.
A 2D-DESI source was mounted onto a SYNAPT G2-Si HDMS Mass Spectrometer. Spray conditions were set as follows: flow rate of 1.5 µL/min, with a 90:10 MeOH:water mixture at 100psi N2 gas pressure, and a voltage of 5kV for both polarities. To conduct the imaging experiment, a raster pattern was defined over the tissue region of interest and the scan speed and line spacing were selected appropriately for the target pixel dimensions. For 150 µm resolution images, the stage was scanned at 0.15 mm per second on the X-axis; and stepped 0.15 mm in the Y-axis between each DESI line scan. In all instances, the MS scan time was 1 second.
As the flow rates used are sufficiently low and the desorption was considered a soft event, the same tissue section can be analyzed more than once without modification or exhaustion of the surface molecules- allowing dual polarity analysis on the exact same section for increased information depth.
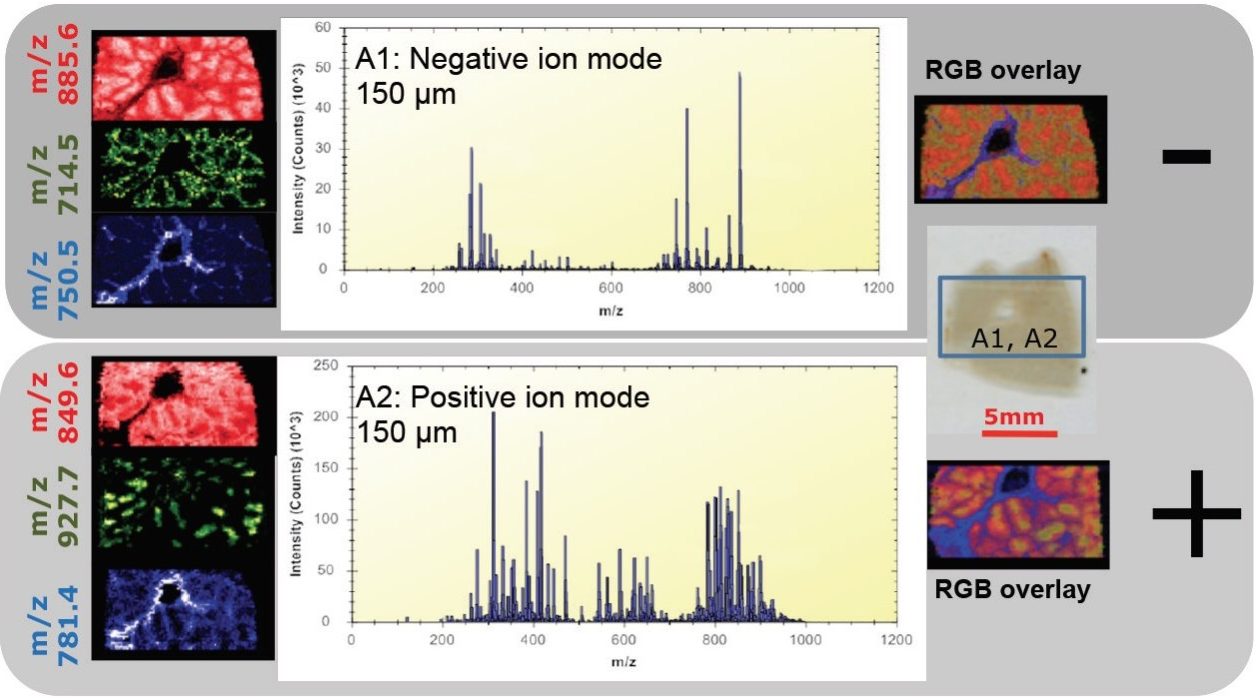
Initially, imaging experiments on porcine liver were performed with the MS operating in negative mode, subsequently followed by imaging the same tissue section in positive mode (Figure 1). In both modes of ionization, plentiful lipids and endogenous metabolites were detected, giving intense peaks for analysis by the mass spectrometer.
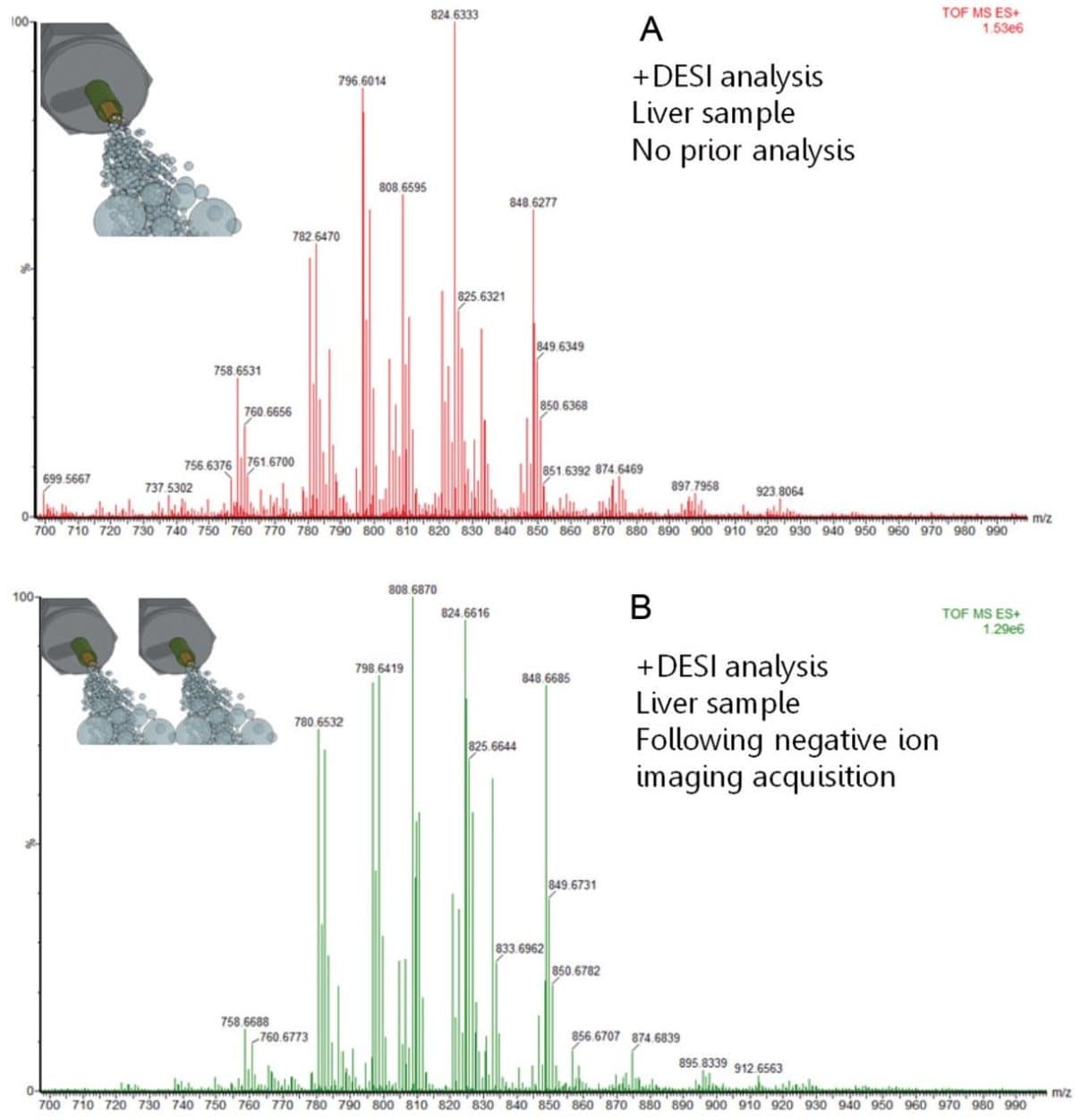
A second experiment was designed to evaluate whether the repeated imaging of the same tissue sample alters the chemical information obtained. Figure 2 compares the spectrum from a single DESI imaging experiment in positive mode (top), with the positive mode spectrum (bottom) generated from a consecutive tissue section, after first analyzing it in negative mode. Identical peaks were observed in both spectra with very similar relative intensities.
The ability to revisit the same section to increase the amount of information could be of great importance when samples are precious, for example with a human liver sample. Figure 3 shows one such example where the same section was analyzed by MS imaging in both polarities. Lipid species specific to tumor tissue and healthy tissue were identified in positive and negative ion mode. This would seem to indicate that the tissue has not been significantly affected by the DESI imaging technique and is still relatively intact. By not perturbing the tissue with DESI imaging, subsequent additional surface analysis or staining techniques (i.e., H&E staining) could be utilized on the same tissue sample for further, more comprehensive characterization.

DESI imaging provides metabolite and lipid molecular information directly from a tissue section with no tissue sample pre-treatment. With low flow rates, the tissue section can be analyzed multiple times without significant degradation of signal or modification of the chemical signature obtained from the tissue. Using the same analytical MS instrument setup, information rich spectra in both positive and negative ion mode can be generated without the need to change solvent or analysis conditions. Therefore, positive and negative ion mode DESI data can be obtained from the same tissue and combined for extended chemical coverage and sample differentiation.
Here we demonstrate that by optimizing DESI imaging conditions, a wealth of molecular information can be accessed from a single tissue section. Multiple images containing unique information can be collected from a tissue sample without the need for the analysis of a number of serial sections.
The advantages of DESI imaging include:
720005299, February 2015