In the work presented here, we show the development and validation of a rapid and sensitive LC-MS method for the simultaneous detection and quantification of multiple, selected CYP450 substrates and metabolites.
High-throughput absorption, distribution, metabolism, and excretion (ADME) screening for the properties of drug candidates have become an essential part of modern drug development. Such screening provides a basis for better information and, consequently, better decision-making in the drug discovery and development process.1 As part of these studies, incubating candidate compounds together with liver microsomes containing cytochrome CYP450 is relied upon extensively to determine compound metabolism, stability, and possible drug-drug interactions (DDI).
The use of CYP450 to determine drug-drug interactions is carried out through inhibition and induction studies and is of particular importance because as the number of prescribed drugs increases, the greater the probability that an adverse drug reaction could exist. Also, it has been stated that there could be up to a 40% probability of a DDI when a patient is administered ten drugs or more.2 In a standard CYP450 inhibition and induction assay, multiple test compounds are evaluated to determine their ability to alter or influence the metabolism of known CYP450 specific substrates.
In the work presented here, we show the development and validation of a rapid and sensitive LC-MS method for the simultaneous detection and quantification of multiple, selected CYP450 substrates and metabolites.

|
LC conditions |
|
|---|---|
|
LC system: |
ACQUITY UPLC I-Class |
|
Vials: |
Waters Maximum Recovery |
|
Column: |
ACQUITY UPLC BEH C18, 3.0 x 50 mm, 1.7 μm |
|
Column temp.: |
35 °C |
|
Sample temp.: |
Ambient |
|
Injection volume: |
5 uL |
|
Flow rate: |
0.600 mL/min |
|
Mobile phase A: |
2 mM NH4CH4COOH, 0.1% NH4OH in H2O |
|
Mobile phase B: |
2 mM NH4CH3COOH, 0.1% NH4OH in CH3OH |
|
Gradient: |
1% to 90% B over 2 minutes; curve 4 |
|
MS system: |
Xevo TQ-S micro |
|
Ionization mode: |
ESI+ |
|
Capillary voltage: |
1 kV |
|
Acquisition mode: |
MRM |
MassLynx Mass Spectrometry Software
The number and diversity of new drug candidates entering the pharmaceutical pipeline has increased, necessitating strategies such as rapid ADME assays for obtaining information that can be used to better qualify candidates. The rapid determination of potential DDIs is of particular importance. If they are discovered late in the development process or after the drug goes to market, DDIs can cost pharmaceutical companies millions of dollars in lost revenue. Figure 2 depicts the separation of two substrates and six metabolites routinely monitored in testing for potential DDIs through inhibition and induction CYP450 assays. Good separation is shown with base widths of chromatographic peaks on the order of three seconds. These results were obtained under generic conditions, illustrating the ease and time for method development.
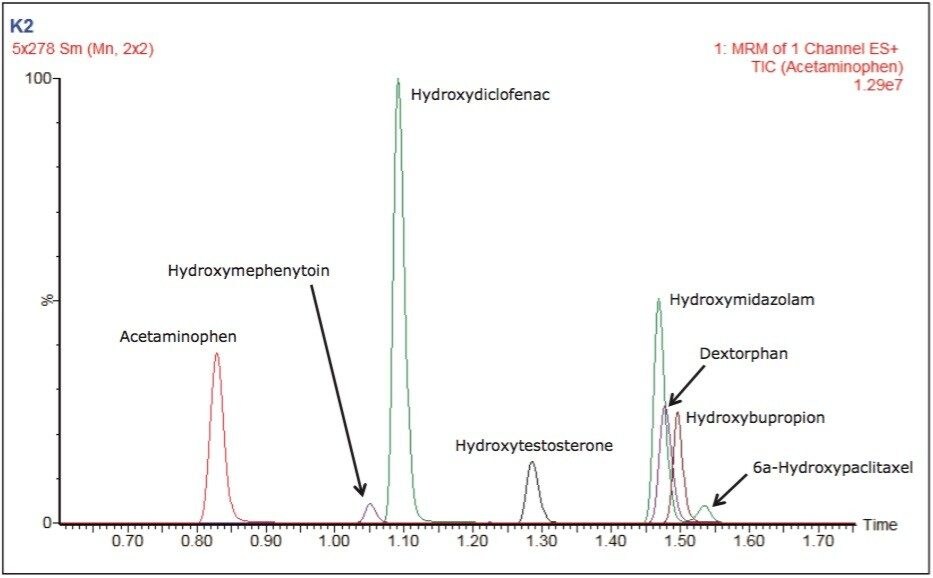
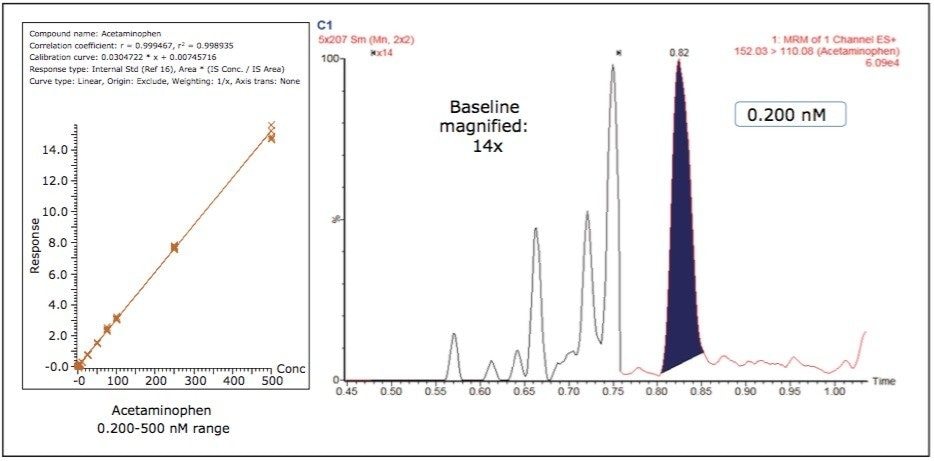
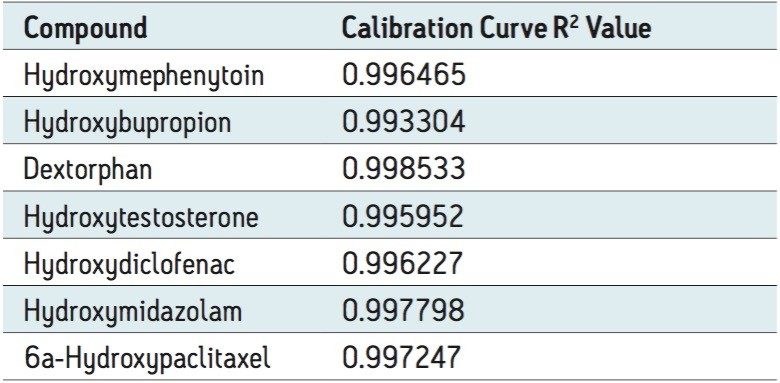
Figure 3 shows the calibration curve for the CYP450 isoform 1A2 substrate acetaminophen. It is linear well beyond three orders of magnitude, and it produces an R2 value of 0.998935 (Table 1 shows the R2 values for the other compounds used in this assay) Figure 3 also shows a lower limit of quantification (LLOQ) of 0.2 nM, with greater than 14 times the signal-to-noise ratio, as measured from the baseline of the separation.
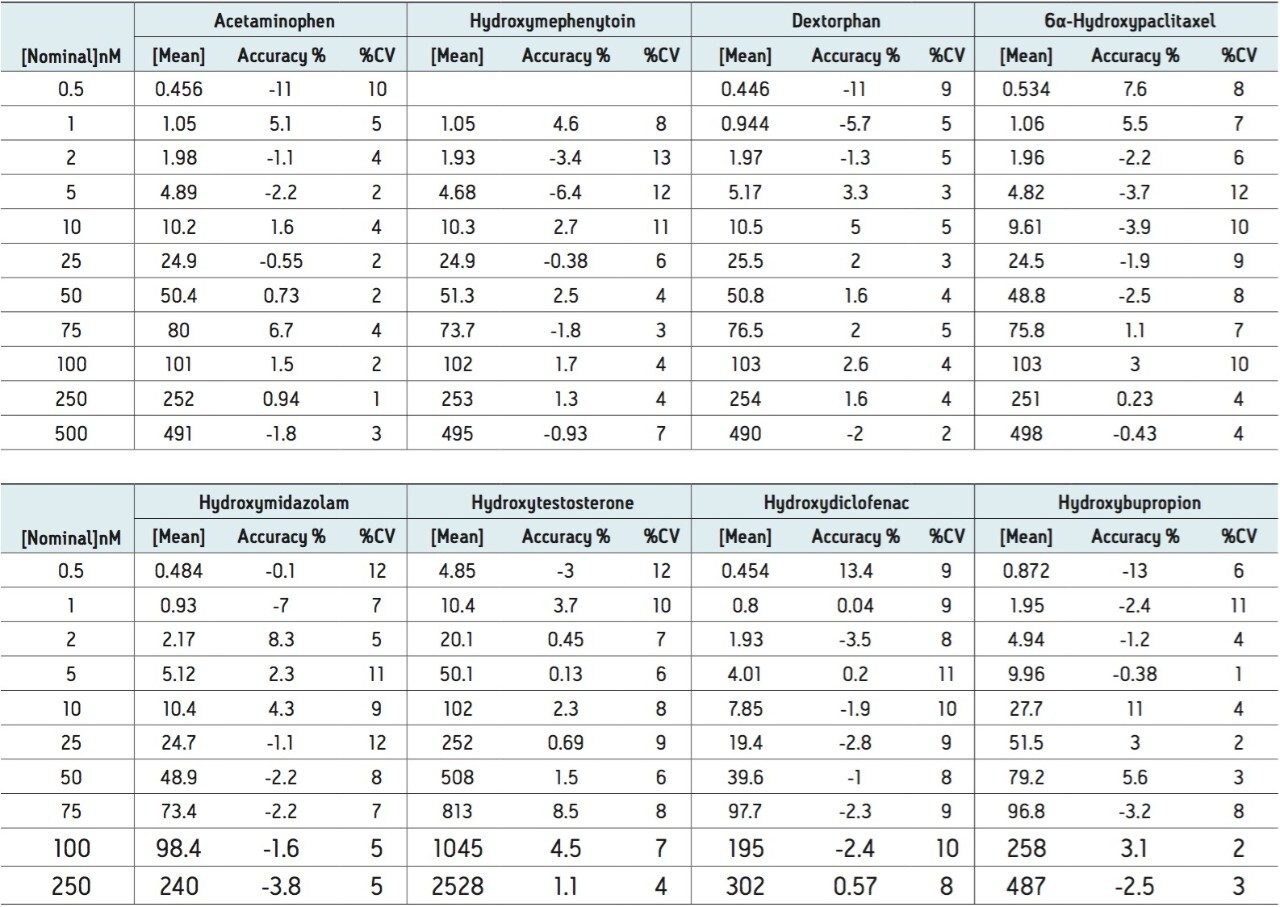
The developed method was then tested for accuracy and precision by LC-MS, as prescribed in the USFDA guidance on bioanalytical method validation. Five determinations were made for 10 concentrations ranging from 0.2 to 5000 nM. The data in Table 2 show that the accuracy and precision for all of the analytes tested falls well within the 15% CV limit (3).
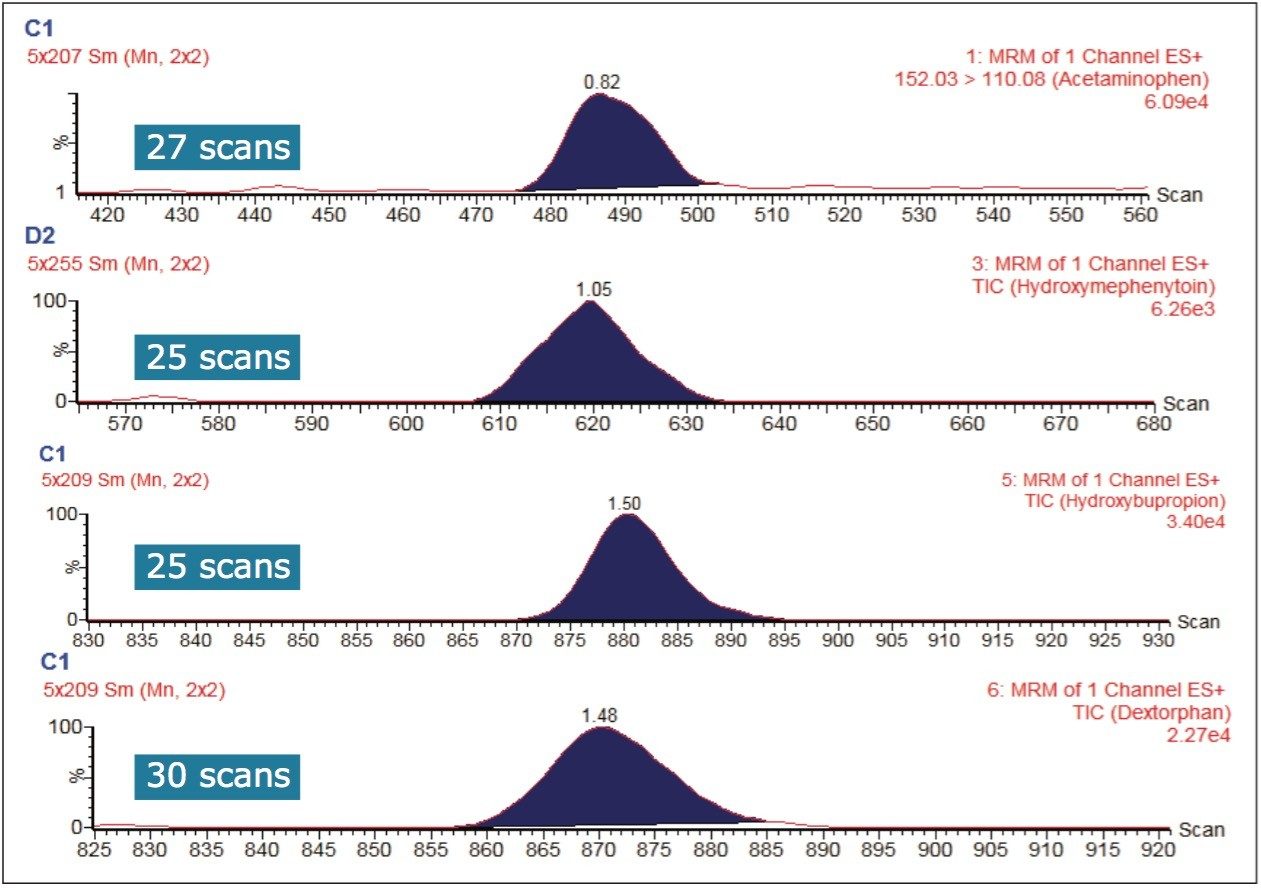
The sensitivity of a bioanalytical LC-MS assay can depend on matrix suppression, chromatographic peak shape, and the duration over which the mass spectrometer samples the chromatographic peak (or peaks) of interest. The sensitivity in this work can also be attributed to the incubation conditions: longer incubation times or higher enzyme concentrations can lead to the development of greater amounts of detected, metabolized substrate. The narrow peak widths produced by the sub-2-μm LC separation require a sufficient number of points across the chromatographic peak for good quantification. Figure 4 shows that the Xevo TQ-S micro is fully capable of acquiring more than 20 points across these narrow peak widths while subsequently monitoring multiple MRM transitions.
The Xevo TQ-S micro can also be operated in RADAR acquisition mode. This allows the simultaneous acquisition of full-scan and MRM data in a single experiment. In Figure 5, we observe the data from the MRM channels monitored in this experiment and also the data from a full MS scan from m/z 50–600. We also observe co-elution of the target analyte with components in the matrix. Thus, the ability to monitor MRM channels and full-scan data can aid in the rapid development of robust methods that minimize the potential for co-elutions and possible matrix suppression. The capability of monitoring full-scan and MRM-channel data could be exploited for drug stability or metabolite identification studies. Though certain chemical entities would lay outside the specific MRM channels, they would nevertheless be subject to detection during full-scan acquisition. Data obtained in this way could then be further verified to determine positive identification.

The combination of the Xevo TQ-S micro and ACQUITY UPLC I-Class produced a rapid, sensitive, and robust method for the separation and detection of multiple, selected CYP450 substrates and metabolites often utilized in DDI studies. The accuracy and precision of the data produced from the method was well within the limits designated by the USFDA guidance for bioanalytical method validation. Linear responses for the calibration curves were obtained, revealing an LLOQ for the analytes of 0.2 nM. Further, the RADAR acquisition mode of the Xevo TQ-S micro enables the acquisition of full-scan and MRM data in a single run; a feature that can aid in method development and other applications.
720005241, December 2014