This is an Application Brief and does not contain a detailed Experimental section.
To demonstrate the ability of the UNIFI Collision Cross Section (CCS) Research Edition to process ion mobility (HDMS) data in a routine, workflow driven manner. Also, to demonstrate the utility of ion mobility to provide orthogonal information, enhancing high resolution MS (HRMS) specificity and allowing complex samples to be understood without complication from unrelated interferences.
Using an orthogonal axis of ion mobility, all interfering and unrelated peaks are removed from the spectra, leaving a spectrum completely free of matrix background.
Metabolite identification requires confident assignment of drug-related components. Whether you are interested in major metabolite pathways to reduce clearance, or concerned with metabolites in safety testing (MIST) guidance, generating fast and informative answers from HRMS data sets is critical. Identifying drug metabolites is a challenging problem. Achieving this in an efficient and accurate manner is an even greater challenge. In biological systems, issues that must be resolved include:
HRMS typically derives specificity through a combination of mass spectral resolution and mass accuracy. Unfortunately, these data alone do not differentiate between isomeric species. Furthermore, understanding and annotating spectra that have interferences can be challenging to the non-expert.
Using an additional chromatographic separation to solve these issues is an attractive proposition. However, traditional 2D LC experiments have limitations in terms of selectivity (for instance, resolving structural isomers) and complexity of method setup. An alternative and attractive approach is to use a technique that separates all components based on their size and shape, as opposed to their electrostatic properties. Drift time and/or CCS can be used to generate this orthogonal, additional dimension of separation of ions via the use of T-Wave ion mobility, and it provides a range of valuable analytical benefits including the ability to generate very clean, or pure, mass spectral data even from samples with a very high matrix burden.
Routine, embedded ion mobility acquisition and processing, measuring the shape and size for all components in the sample is now significantly enhanced by the SYNAPT G2-Si platform. With this hardware and software solution, we can generate significantly cleaner chromatograms and MS spectra, thereby reducing analytical complexity. In addition to distinguishing the biological matrix from the analyte based on the size and shape, this approach also allows structural isomersto be distinguished on the basis of differences in shape. UNIFI then provides users with clean and highly selective data (mass, retention time, peak intensity, and now size and shape) in a simple workflow-driven software package.
Propranolol standard curves were spiked into plasma, and protein-precipitated using two volumes of acetonitrile to one volume plasma. 5 μL of the supernatant was injected onto the LC/MS system using a ballistic gradient (5% to 95% over two minutes). Propranolol eluted at a retention time of 1.1 minutes. Mass spectrometry was carried out on a SYNAPT G2-Si and data collected in mobility resolution mode. Data was collected using HDMSE mode (full scan m/z 50 to 1000 for precursors and product ions along with full ion mobility characterization) controlled by MassLynx Software. Sample data was imported into UNIFI CCS Research Edition, and processed using a standard Metabolite Identification workflow.
Figure 1 shows the raw full scan spectrum of propranolol at a concentration of 0.12 ng/mL. At these concentrations, the ion for the parent compound is completely hidden by the complex background matrix found in plasma. Even at zoomed inset (200 to 300 m/z range), the parent ion is still obscured by the wide variety of other ions eluting at the same retention time.
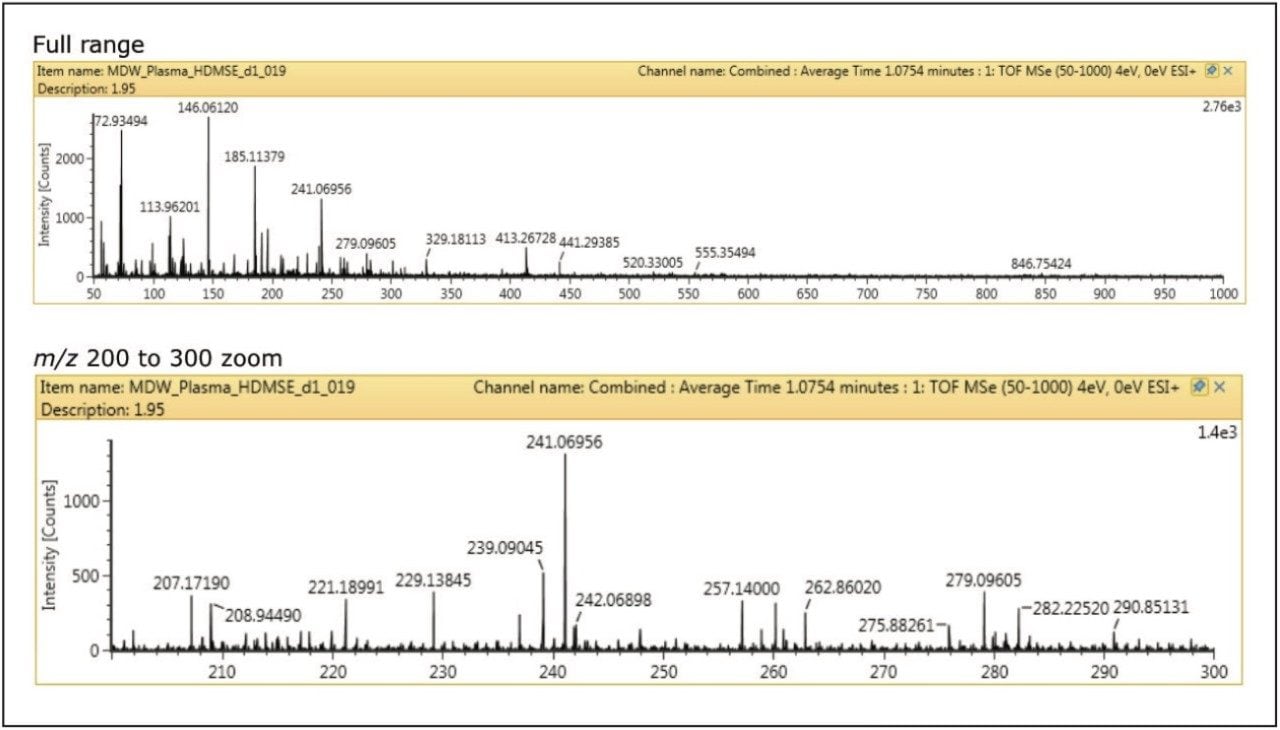
Figure 2 shows the first step of data cleanup. This image shows data that have been time-aligned and cleaned up, but without the extra dimension of separation by size/shape. Most of the interference is removed allowing for much simpler identification of the parent compound from the spectral data.
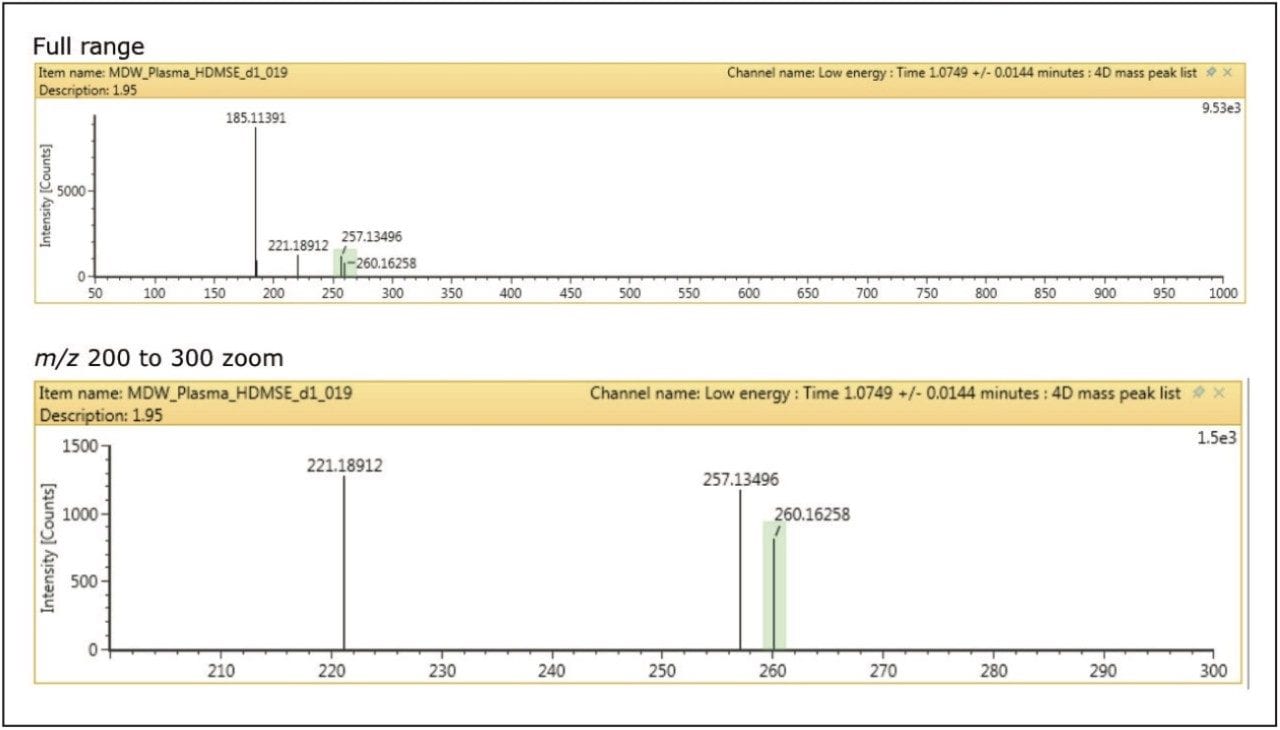
Figure 3 shows data that has also been cleaned up using the orthogonal axis of ion mobility. All interfering and unrelated peaks are removed from the spectra, leaving a spectrum completely free of matrix background. This additional orthogonal separation makes the HRMS identification simple and easy to interpret.
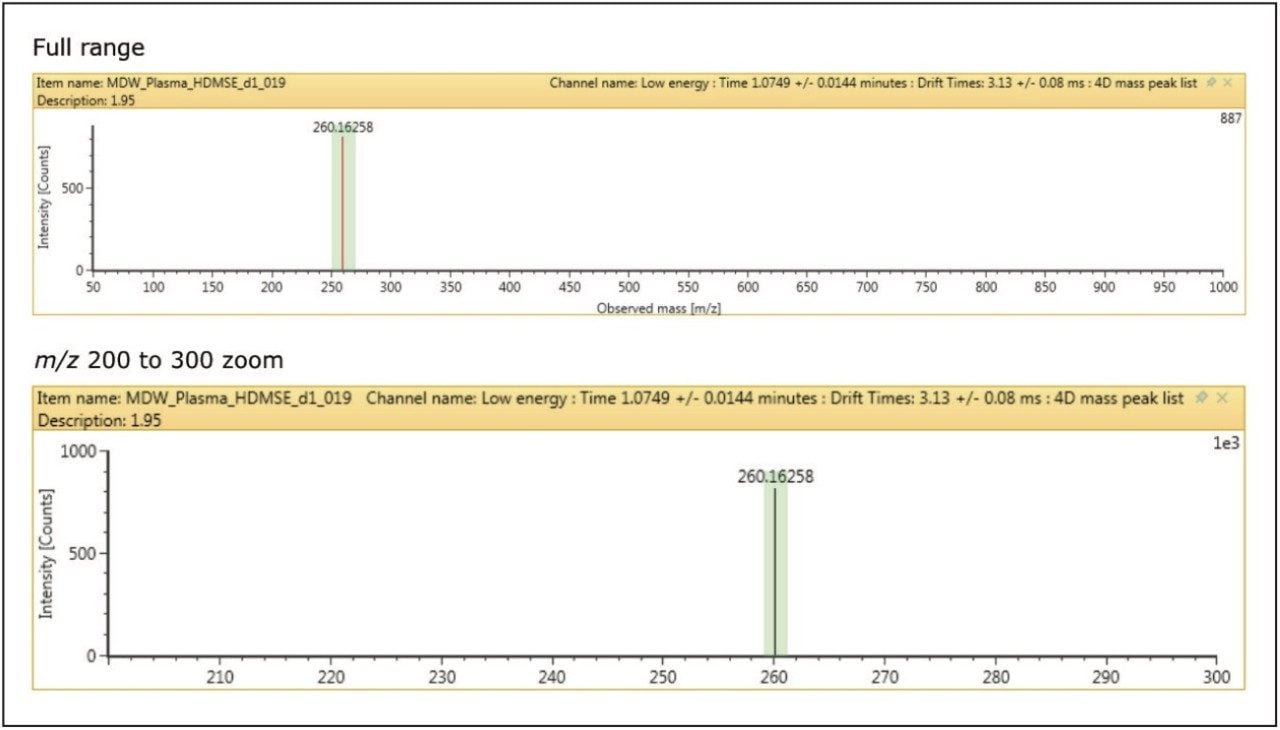
Orthogonal size/shape separation is a simple and powerful technique embedded in the SYNAPT G2-Si. It provides higher specificity and cleaner data than is possible with any other HRMS technique.
The enhancements available by processing this data with UNIFI CCS Research Edition software give the scientist access to the added benefits of orthogonal size/shape separation without changes to current workflows, allowing metabolite identification challenges to be addressed with speed and confidence.
720004741, June 2013