The Waters UNIFI Scientific Information System employs a workflow approach with acquisition templates and promotable parameters to enable the rapid simple screening and optimization of gradient and column temperature conditions. Further optimization of the chromatographic parameters can significantly improve assay robustness and analytical throughput.
Maximize bioanalytical assay productivity and throughput using promoted parameters and template acquisition with the Waters UNIFI Scientific Information System.
Developing and optimizing the chromatographic conditions for a bioanalytical method is a multi-step process that includes screening column chemistries, mobile phase organic modifier, mobile phase pH, and column temperature. This entails not only the evaluation of the basic chromatographic performance of the system (retention time, peak shape, tailing factor, etc.), but also resolution of the target analyte from endogenous components in the matrix.
After the mobile phase solvents and column chemistry have been selected, the gradient steepness, starting concentration and final composition can be optimized to maximize analyte resolution and minimize the analytical run time to improve sample throughput.
Screening the various gradient conditions to select the best conditions requires multiple experimental runs and careful review and comparison of the elution characteristic of the target analyte(s) from the endogenous components in the sample. Previously we have described the use of functionality of promoted parameters and templated data acquisition within the UNIFI Scientific Information System to streamline the acquisition of LC-MS data and thus simplify the method development process.

|
LC system: |
Waters ACQUITY UPLC |
|
LC column: |
ACQUITY UPLC BEH C18 1.7-μm, 2.1 x 50 mm |
|
MS system: |
Waters Xevo TQ-S |
The separations were performed using a mobile phase of 0.1% (v/v) formic acid and acetonitrile as the organic modifier
The gradient steepness and column temperature were was adjusted as a variable during the analytical acquisition process.
The mass spectrometer was operated in electrospray positive ion mode with MRM data collection, RADAR full scan data collection and precursors of m/z = 184 to monitor the phospholipid elution profile.
Biological samples were prepared by spiking the target analyte into control human plasma. The spiked sample was then precipitated using a 2:1 ratio of acetonitrile to plasma. The resulting sample was centrifuged at 13,000 RPM and the supernatant removed for analysis by LC-MS.
The acquisition of data within the Waters UNIFI Scientific Information System is workflow-driven, allowing scientific managers to develop and administer templates for their scientist to employ for multiple tasks, including routine batch analysis, matrix effects calculation, gradient screening and gradient optimization. This use of a workflow-driven approach ensures consistency and allows the less experienced scientist a structured route to analytical method development and optimization.
Within the UNIFI Scientific Information System, the user is presented with a series of workflows to select from, Figure 2.
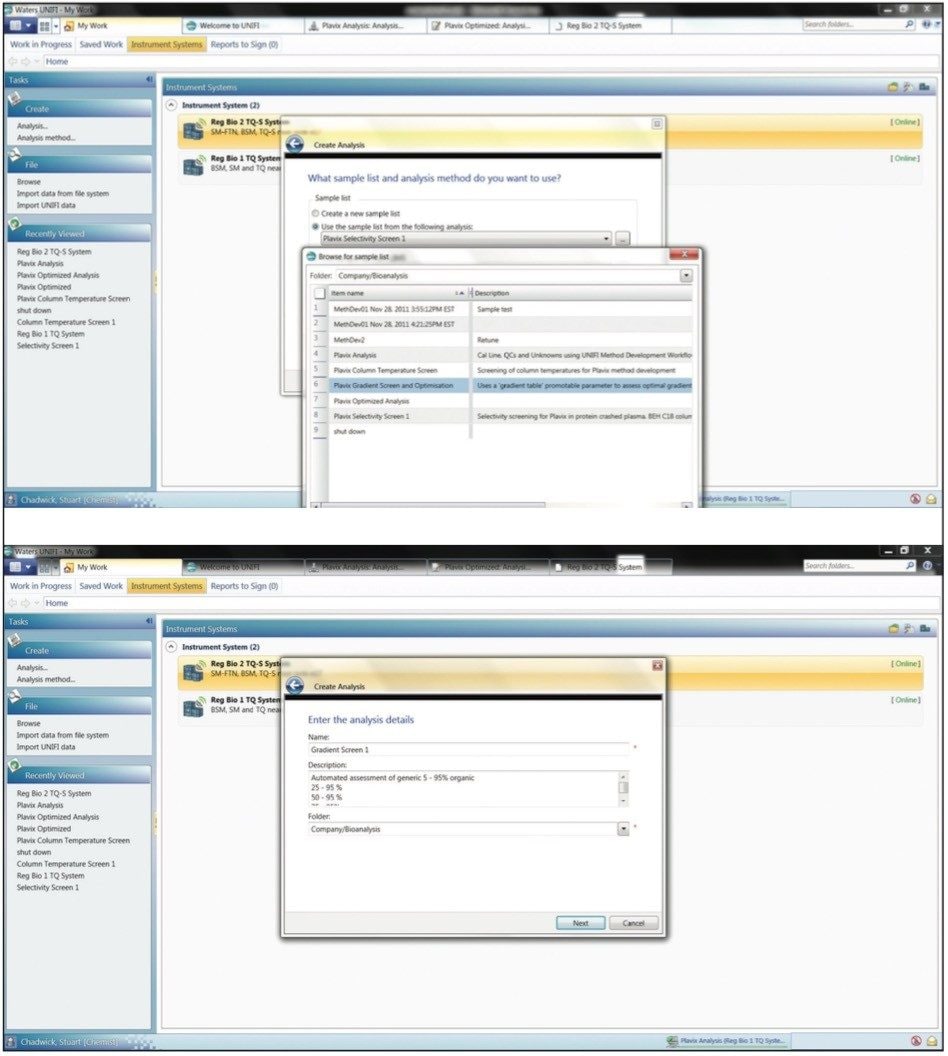
After selection of the analysis method and sample list to be used, the user then saves the analysis to the appropriate folder. The scientist can then select the different gradient steepness and shapes to be evaluated via a drop-down menu, as shown in Figure 3. This prevents the propagation of numerous methods, by different scientists and ensures consistent operation.
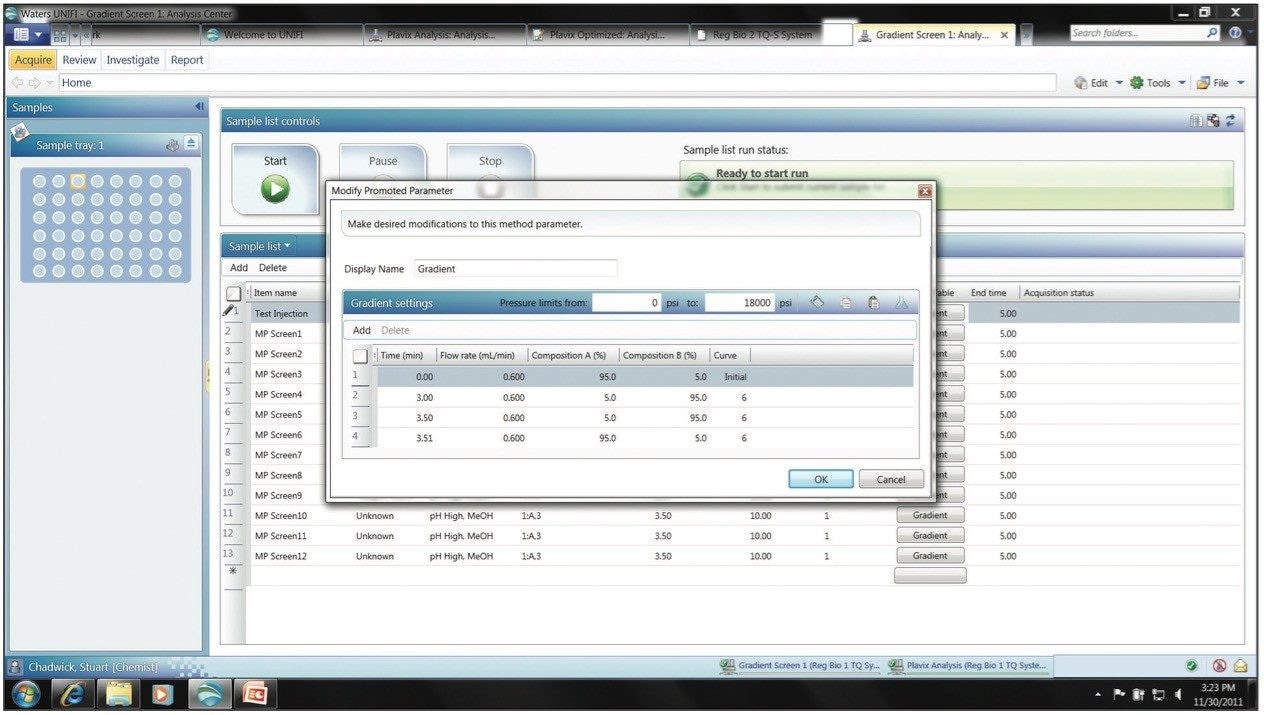
Once the various gradient slopes to be evaluated have been selected the acquisition can be started and the data acquired. Following data acquisition the data can be reviewed to select not only the system that will allow the greatest throughput, but also that which will deliver the greatest robustness.
In this case four different gradient slopes were evaluated: A 5-95%; B 5-50%; C 25-75%; and D 50-75%. The evaluation of the resolution and effectiveness of the different gradient shapes was performed by comparing the retention time of the target analytes with that of the endogenous components in the sample. The endogenous components were monitored by the simultaneous acquisition of full scan MS data and precursors of m/z = 184 to track the elution profile of the phospholipids. The data displayed in Figure 4 shows the elution profile of the target compound and the endogenous components in the sample
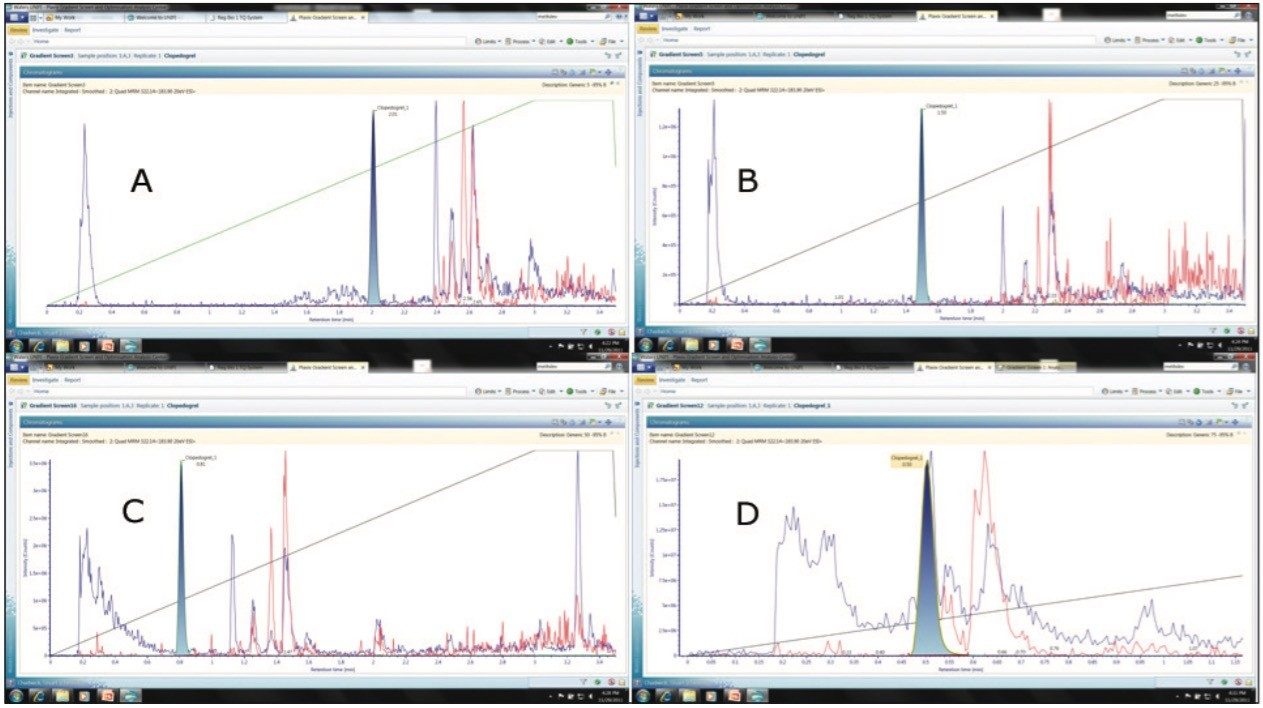
We can see from this data that composition A gave clear resolution of the analytes from the endogenous components as did gradients B and C, with gradient C delivering the shortest analysis time of the three. Gradient D had the greatest throughput of the four, however the analyte peak eluted in the same region of the chromatogram as the endogenous components. Thus for this assay gradient C would be the best option, delivering the best compromise of throughput and resolution. By employing gradient C for the assay the compound elution time has been reduced from 2.01 minutes to 0.81 minutes allowing the LC condition to be modified to effectively double the throughput.
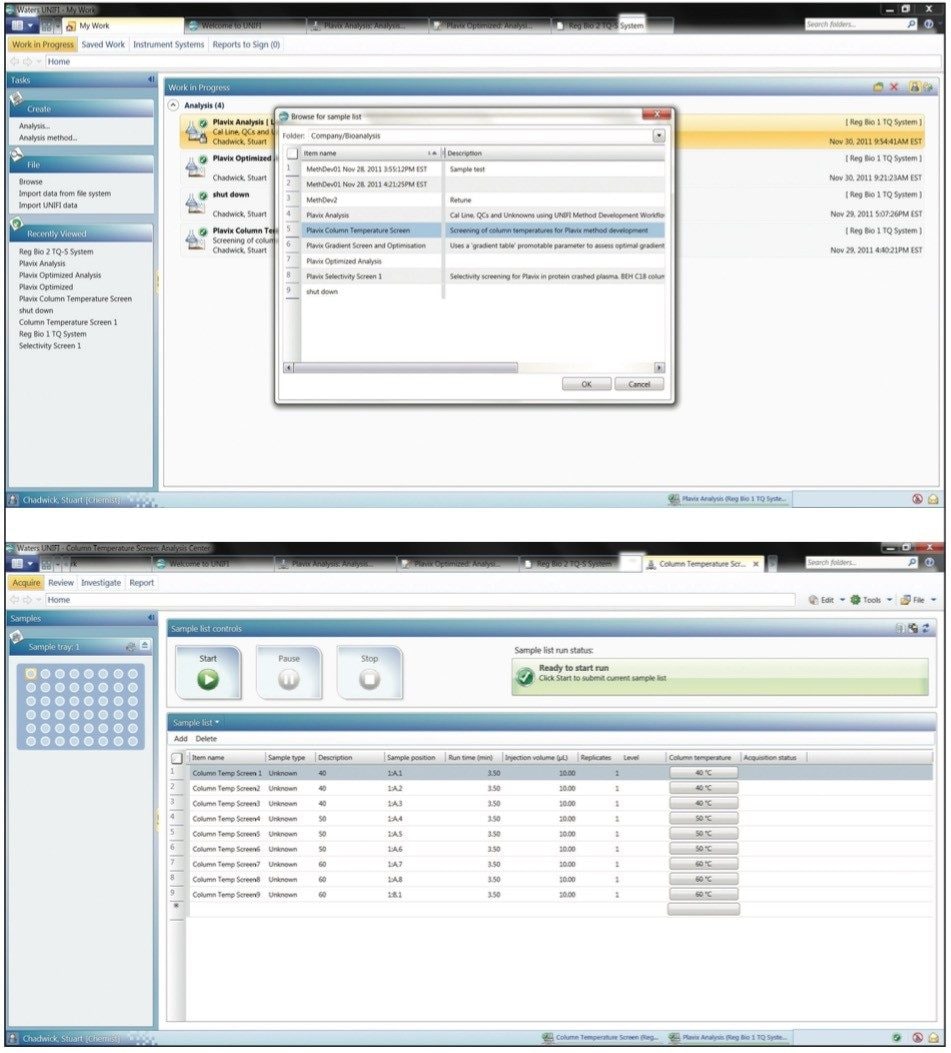
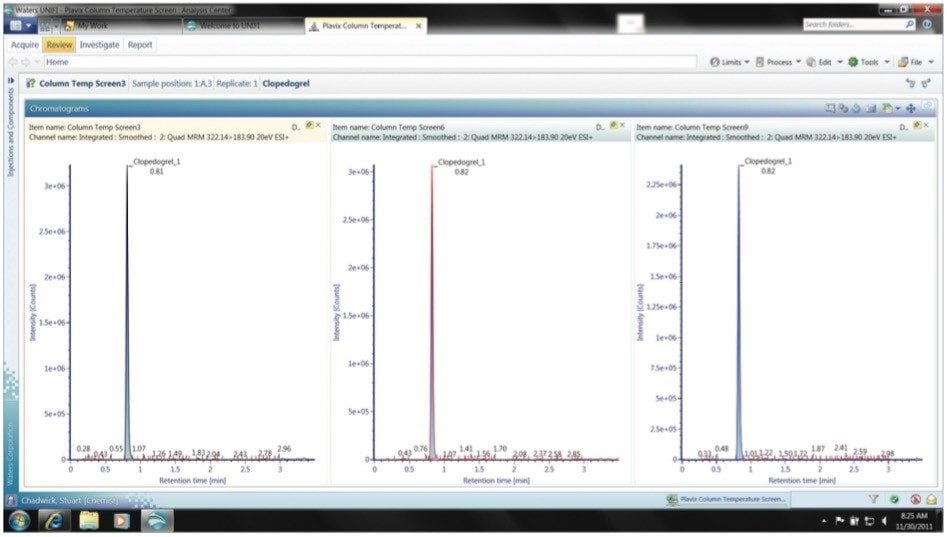
Chromatographic column temperature can be evaluated in a similar manner to that employed for gradient steepness and slope for method development. Using a similar template approach, the column temperature can be varied, Figure 5. In this case column temperatures of 40 °C, 50 °C, and 60 °C were evaluated. The results generated showed that, in this case, little benefit is gained in terms of peak shape or retention time when the column temperature is varied from 40 to 60 °C, Figure 6.
The Waters UNIFI Scientific Information System employs a workflow approach with acquisition templates and promotable parameters to enable the rapid simple screening and optimization of gradient and column temperature conditions. Further optimization of the chromatographic parameters can significantly improve assay robustness and analytical throughput. This workflow approach delivers:
720004288, February 2012