
This application note demonstrates the use of a high sensitivity exact mass time-of-flight (Tof) instrument, the Waters Xevo G2 Tof, for the rapid quantitative/qualitative analysis of pharmaceutical compounds in a drug discovery environment.
The combined qualitative and quantitative capabilities of the Xevo G2 Tof makes it an ideal tool for early drug discovery metabolism studies, providing:
Quantitative bioanalysis support of drug discovery projects has traditionally been performed on tandem quadrupole mass spectrometers coupled to liquid chromatography operated in MRM mode; this is due mainly to the specificity, linearity of response and sensitivity of the instrumentation.1 In contrast, accurate mass MS instrumentation has been reserved for qualitative data analysis such as metabolite identification.2 However, if analyte quantification can be successfully achieved on such an instrument, the latent full-scan data can be of significant use in understanding the reasons behind issues such as poor exposure or rapid compound elimination.
With the drive to improve the efficiency of the drug discovery process many scientists are looking to combine the collection of quantitative and qualitative data into one process.3 In this application note, we present the use of a high sensitivity exact mass time-of-flight (Tof) instrument, the Waters Xevo G2 Tof, for the rapid quantitative/qualitative analysis of pharmaceutical compounds in a drug discovery environment.
The samples, calibration line, and QCs were prepared by spiking authentic standards of the drug substances in solution into rat plasma over the range of 0.1 to 100 ng/mL. The samples were prepared by the protein precipitation of a 50 μL aliquot of plasma with 100 μL of acetonitrile. The samples were centrifuged and the resulting supernatant removed, diluted 1:1 with H2O, and submitted for analysis by UPLC-MSE.
The chromatography was performed on a 2.1 x 50 mm ACQUITY UPLC BEH C18, 1.7-μm column and eluted under reversed phase gradient conditions over 5 minutes using an ACQUITY UPLC System. The column effluent was monitored by electrospray MS operating in positive ion mode on a Xevo G2 Tof System. The data was collected in MSE mode over the mass range of 50 to 1200 m/z. The collision energies, capillary voltage, and cone voltage were set at 6/21 (low/high) eV, 0.5 kV, and 30 V respectively.
In tandem quadrupole mass spectrometry, assay specificity and sensitivity is derived from the use of MRM detection. In this mode of operation only analytes that have a specific precursor mass and produce a defined fragment ion will be detect and measured. With exact mass LC-MS analysis, the specificity of the assay is derived from the ability of the instrument to resolve ions to a high degree of accuracy (three to four decimal places) in the 2 to 5 ppm mass range. In this approach, a narrow mass window for ion extraction is placed around the exact mass of the ion (M+H+) of interest. The Xevo G2 Tof is a high resolution –greater than 20,000 Full Width at Half Maximum (FWHM – orthogonal time-of-flight mass spectrometer delivering mass accuracy of < 2 ppm RMS on complex samples such as those analysed here.
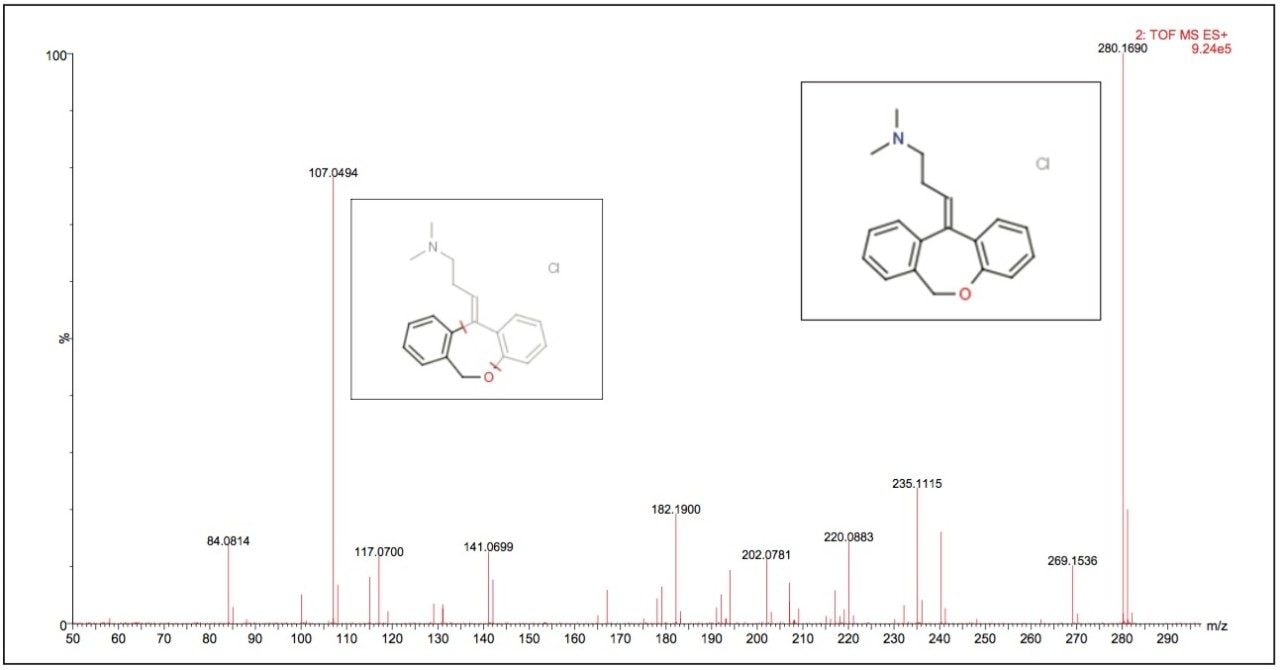
Data displayed in Figure 1 illustrates the MS spectrum of the psychotropic agent doxepin. This compound exhibits both tricyclic antidepressant and anxiolytic properties. Doxepin has a molecular formula of C19H21NO, giving a M+H+ exact mass of 280.1701. The Xevo G2 Tof measured mass of 280.1703 that produced an elemental composition of C19H22NO for the M+H+ ion with a mass error of 0.7 ppm.

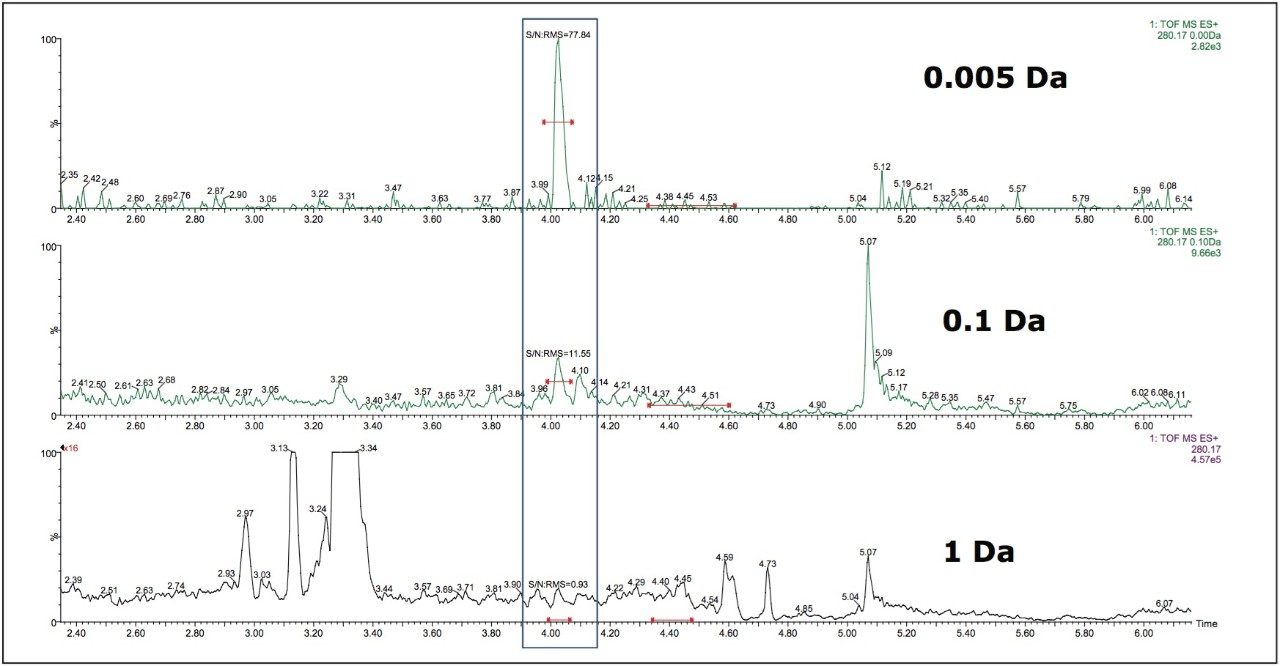
Data in Figure 2 shows the effect on the selectivity and sensitivity of the assay, measured as signal to noise, as the mass window for ion extraction is reduced. In this example, a 50 pg/mL extract of doxepin in rat plasma was analysed by UPLC-MS using a simple organic/aqueous gradient. In the bottom chromatogram a 1 Da window is employed and the doxepin peak, eluting at 4.05 minutes is not visible. If the mass window is reduced to 0.1 Da, centre chromatogram, the peak is just visible with a signal-to-noise value of 11:1. However, as the mass window is reduced to 0.005 Da, top chromatogram, the analyte peak is now clearly visible with a signal-to-noise value of 77:1.
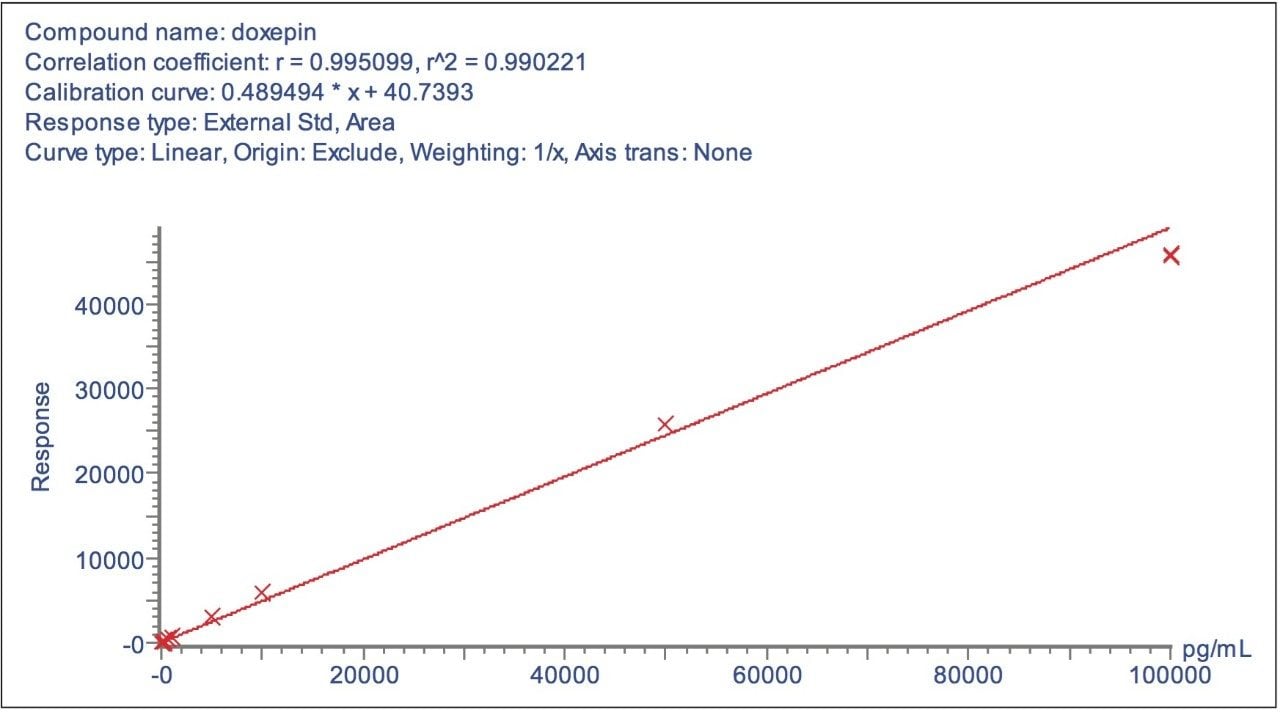
Using this approach the assay was found to be linear over the range of 0.1 to 100 ng/mL, as shown in Figure 3. The ability to use these narrow mass extraction windows is made possible by the mass stability of the QuanTof analyser on the Xevo G2 Tof.
Although the use of exact mass and a narrow mass extraction window allows the quantification of drug analytes in biological fluids, the assay accuracy may still be affected by the coelution of endogenous components in the sample. This issue is addressed in tandem quadrupole mass spectrometry by the use of fragment ions to increase specificity.
The Xevo G2 Tof has the capability of simultaneously acquiring both precursor and product ion information. This is achieved by operating the collision cell in the mass spectrometer at alternating high and low collision energies. However, unlike classical MS/MS, there is no pre-selection of precursor ions so all of the fragments ions generated from all of the ions entering the source are detected and stored. The precursor and product ions are stored in two separate data channels. This mode of operation is called MSE. This is in effect two simultaneous full-scan exact mass experiments.
The advantage in operating the instrument and collecting the data in this manner is that all of the information is stored without need for prior selection or decision-making. This allows the data to be reprocessed and reinterrogated post acquisition without the need for further injections and instrument analysis time to generate product ion spectra on peaks of interest.
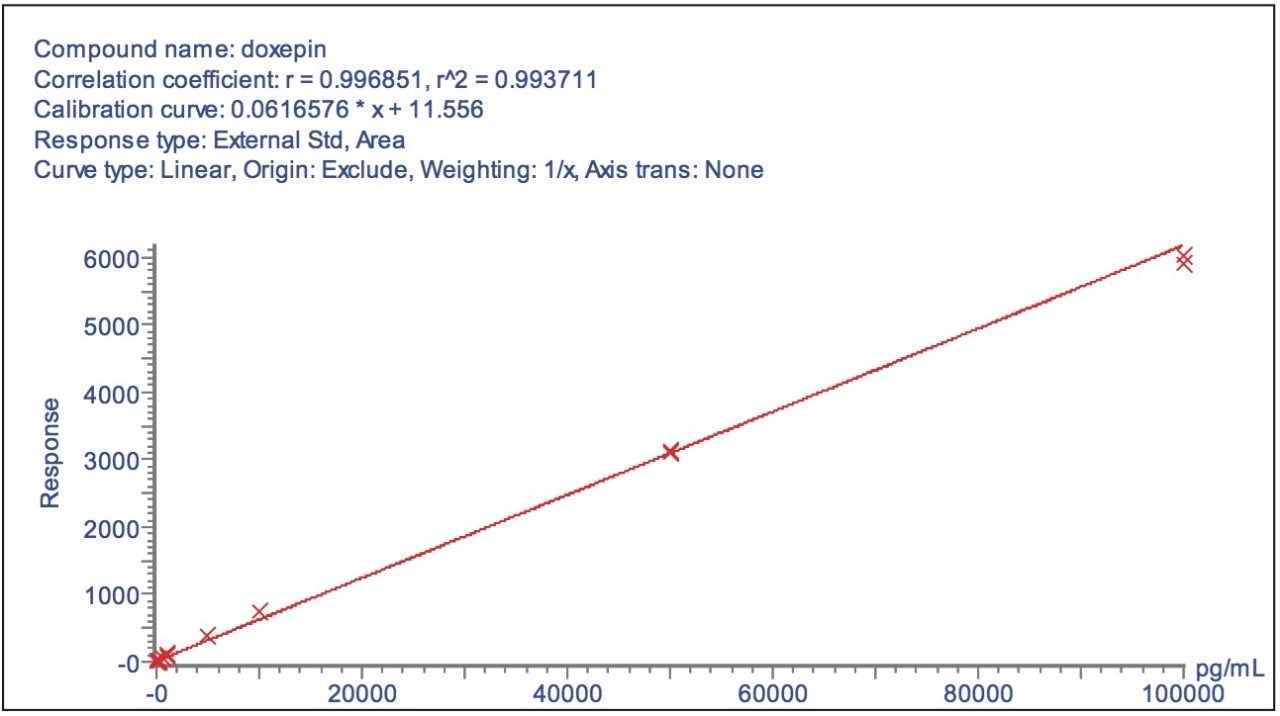
The fragment ion spectrum produced from the high collision energy analysis of doxepin is shown in Figure 4. Here we can see that the major fragment ion produced was m/z 107.0494, the structure of both the parent and fragment ions are displayed in the spectrum. To illustrate the versatility of this approach, a calibration line was created using the m/z 107.0494 product ion from the elevated collision energy data, Figure 5. The assay was again found to be linear over the calibration range of 0.01 ng/mL to 100 ng/mL.
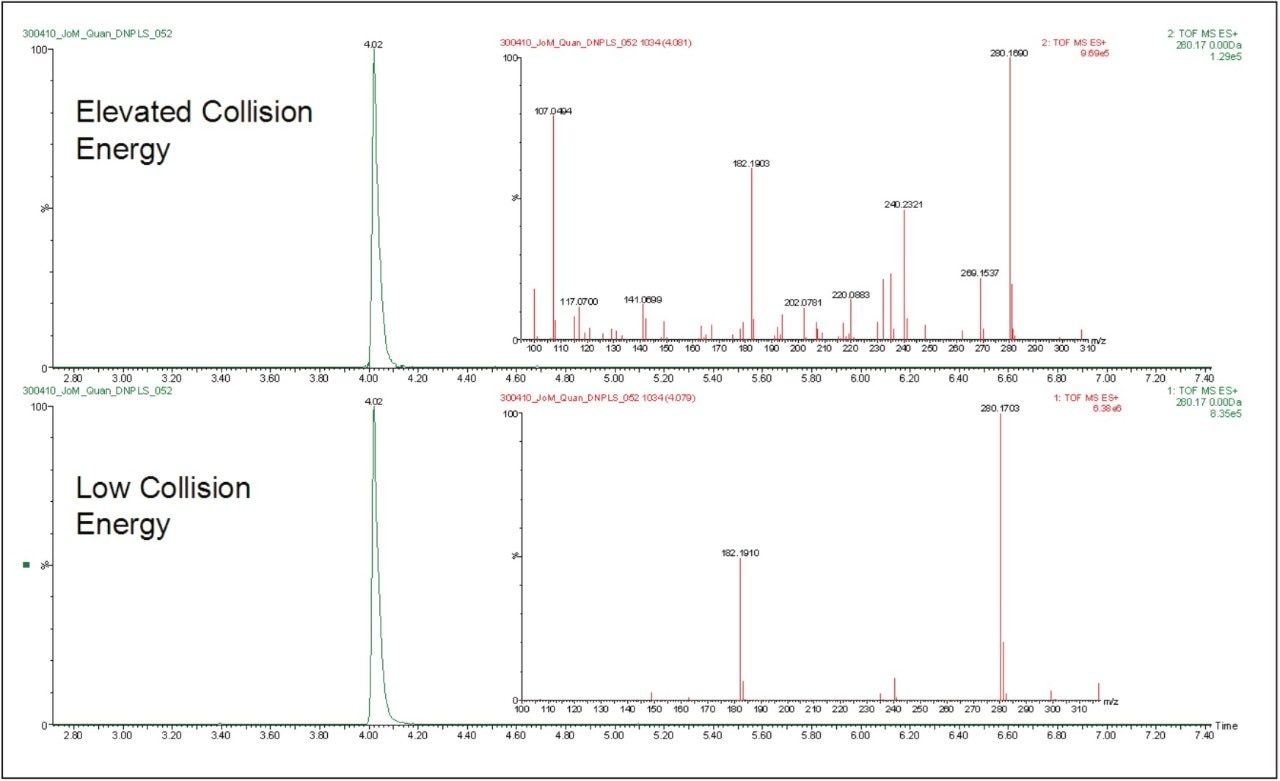
Another major benefit of this approach is that the data is collected and stored in an unbiased manner. Hence, the resulting data contains all of the precursor and product ion information in an exact mass format for future interrogation. This MSE data can be used to confirm the identity of the peaks detected and quantified hence providing further confidence on the identity of the peak in question. The data displayed in Figure 6 show the elevated and low collision energy data from the analysis of a 50 ng/mL standard of doxepin. This latent information makes the confirmation of analyte identity quick and easy as it provides both exact mass precursor and exact mass product ion data.
During a drug discovery project it is often necessary to identify the drug metabolites produced from the biotransformation of the dosed compound. When using a tandem quadrupole MS this often requires further LC-MS experimentation. As previously mentioned the Xevo G2 Tof, when operated in MSE mode, acquires both precursor and product ion data for all of the analytes in the sample in one analytical run. This facilitates the post acquisition mining of the LC-MSE data for drug related metabolites.
This can be accomplished by employing several different tactics. The low collision energy data (precursor ions) can be screened for phase I metabolic additions and cleavages as well as phase II conjugations reactions. The elevated collision energy (product ion) MSE data can be used to manually search for drug related products using a common fragment ion approach. The low and elevated collision data can also be interrogated to generate pseudo-constant neutral loss information to search for potentially toxic metabolites such as glutathiones. The specificity and accuracy of the metabolite detection is enhanced as both modes of acquisition (low and elevated collision energy) are collected in exact mass mode.
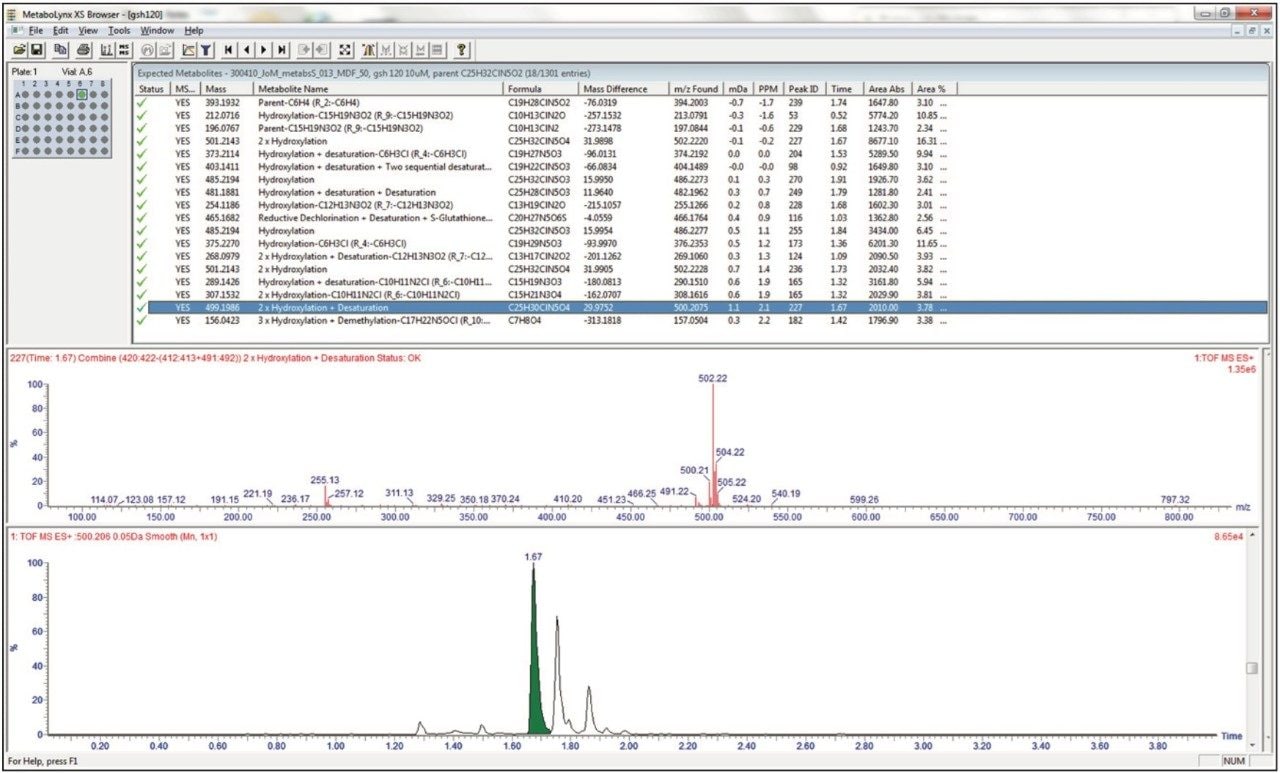
The analysis of the metabolite data is automated, simplified and streamlined by the use of the MetaboLynx XS Application Manager. The data shown below in Figure 7 illustrates the LC-MSE analysis of an in vitro incubation of nefazodone at the 1 μmol level. The results from the MetaboLynx browser are shown with the detected metabolites, elemental composition, mass error and retention time listed in the results table. The browser also allows for the MS spectra and chromatogram to be dynamically searched and evaluated. These data show that 17 different drug metabolites were detected with the results for a 2 x hydroxylation + desaturation metabolite being shown.
Use of full-scan exact mass LC-MS for the quantification of pharmaceutical compounds in biological fluids facilitates fast simple analysis ideal for drug discovery. The high mass accuracy stability derived from the QuanTof mass analyzer, allowing narrow mass extraction windows, confers a high degree of specificity on the assay. The ability of Xevo G2 Tof to simultaneously collect elevated and low collision energy data with a high degree of mass accuracy allows quantification using both precursor and product ions as well as confirmation of analyte peak structure and metabolite identification.
This combination of qualitative and quantitative capability of the Xevo G2 Tof makes it an ideal tool for early drug discovery metabolism studies. With MSE and exact mass UPLC-MS, a laboratory will maximize quality information from samples to achieve superior profiling and identification in less time for the most complex of small molecule analysis.
720003813, November 2010