
Protein phosphorylation is one of the most common post-translational modification occurring in mammalian systems and is especially important in a number of key biological processes, such as intracellular signalling, facilitating the method of action of several key hormones.
Numerous strategies for studying phosphorylation have been described; several incorporating trypsin digestion with subsequent analysis by LC-MS and MS/MS. However, the analysis of small hydrophilic phosphopeptides by LC-MS/MS techniques is particularly challenging due to the poor retention characteristics of these phosphopeptides on reverse phase media. During the HPLC experiment they are often ‘lost’, either by not binding to trapping columns during the loading step often employed in nano LC experiments or by eluting in the void volume for direct loading LC experiments. In addition the analysis of these species by either Electrospray (ESI) or Matrix Assisted Laser Desorption Ionization Mass Spectrometry (MALDI-MS) is complicated by their low molecular weight.
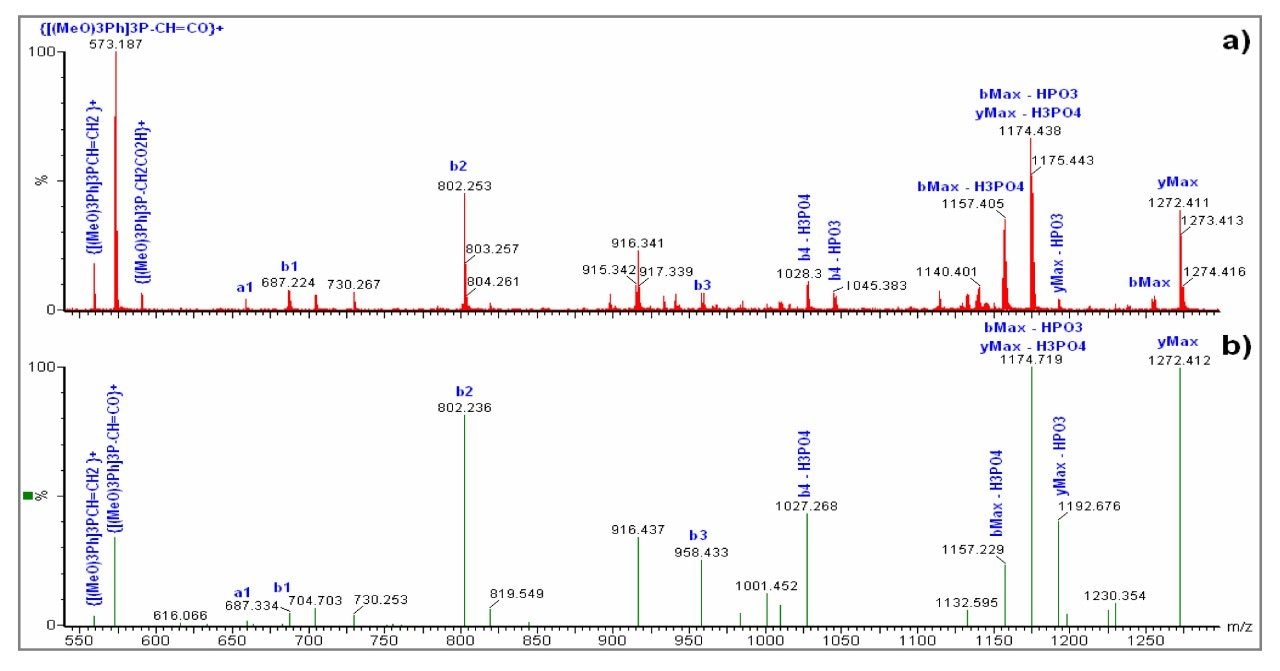
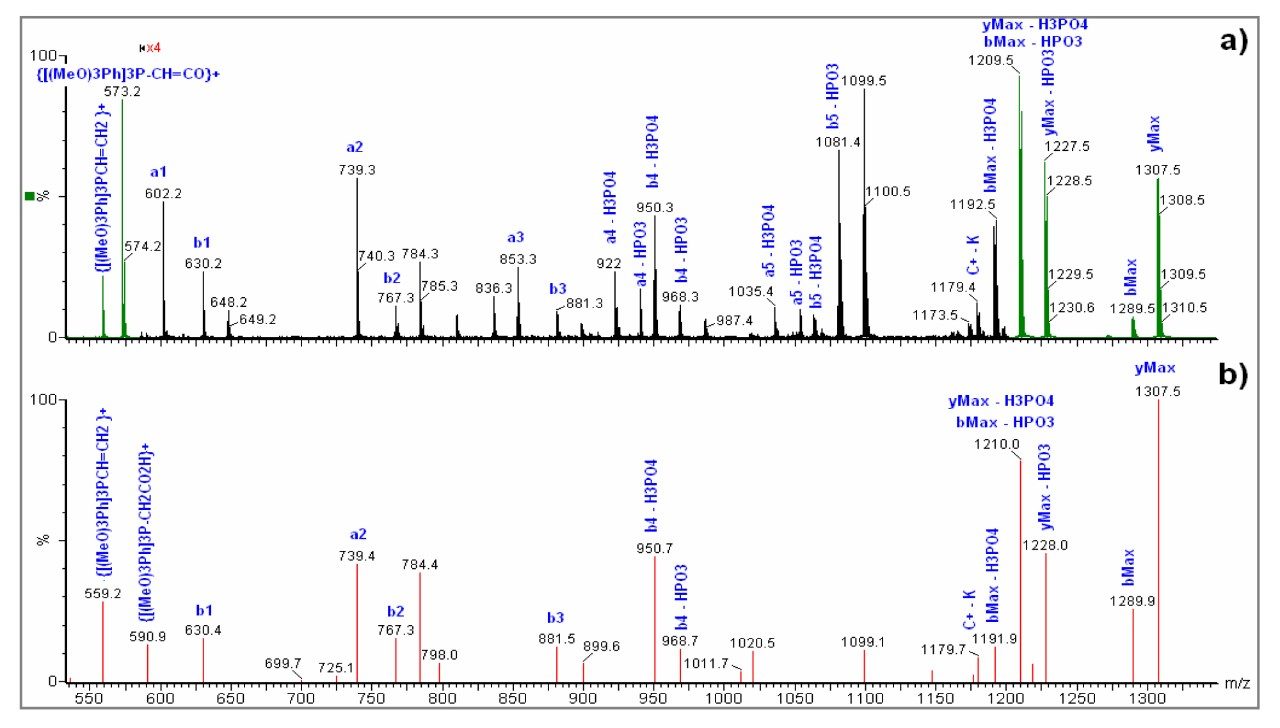
In this work we describe the N-terminal derivatization of small phosphopetides using N-Tris (2,4,6-trimethoxyphenyl) phosphonium-acetic acid N-hydroxysuccinimide ester (TMPP-ac-OSu). This derivatization chemistry improves phosphopeptide retention on reverse phase materials and increases the mass of peptides by 572 Da whilst also enhancing fragmentation in the MS/MS mode for singly charged peptides.1,2

NDRSpE (MW 699.2225 Da)
DSpT (MW 401.0835 Da)
GHNSpLK (MW 734.3113 Da)
SNEDYpR (MW 862.2858 Da)
Phosphopeptide samples were analyzed on the QTof Premier using nanoscale LC-MS. Chromatographic separation was achieved using a Waters NanoEase Atlantis dC18 (100 mm x 75 μM) analytical column, using a gradient from 5% to 85% acetonitrile over 45 minutes.
125 fmol of each TMPP modified phosphopeptide were injected.
All data was acquired on the QTof Premier in continuum mode over the m/z ranges 50-1990 in the W-Optics mode of operation.
Instrument resolution was better than 17,500 FWHM.
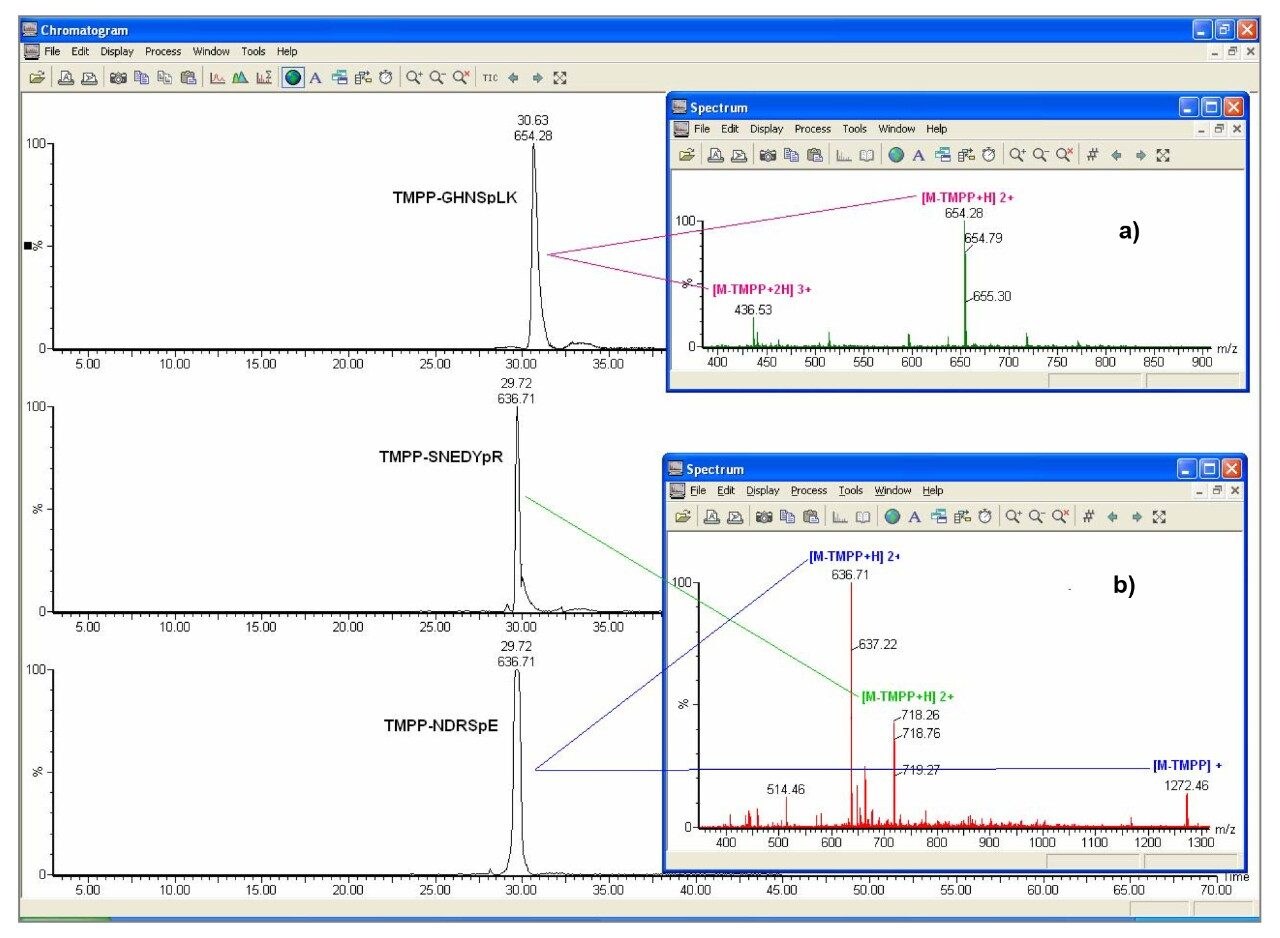
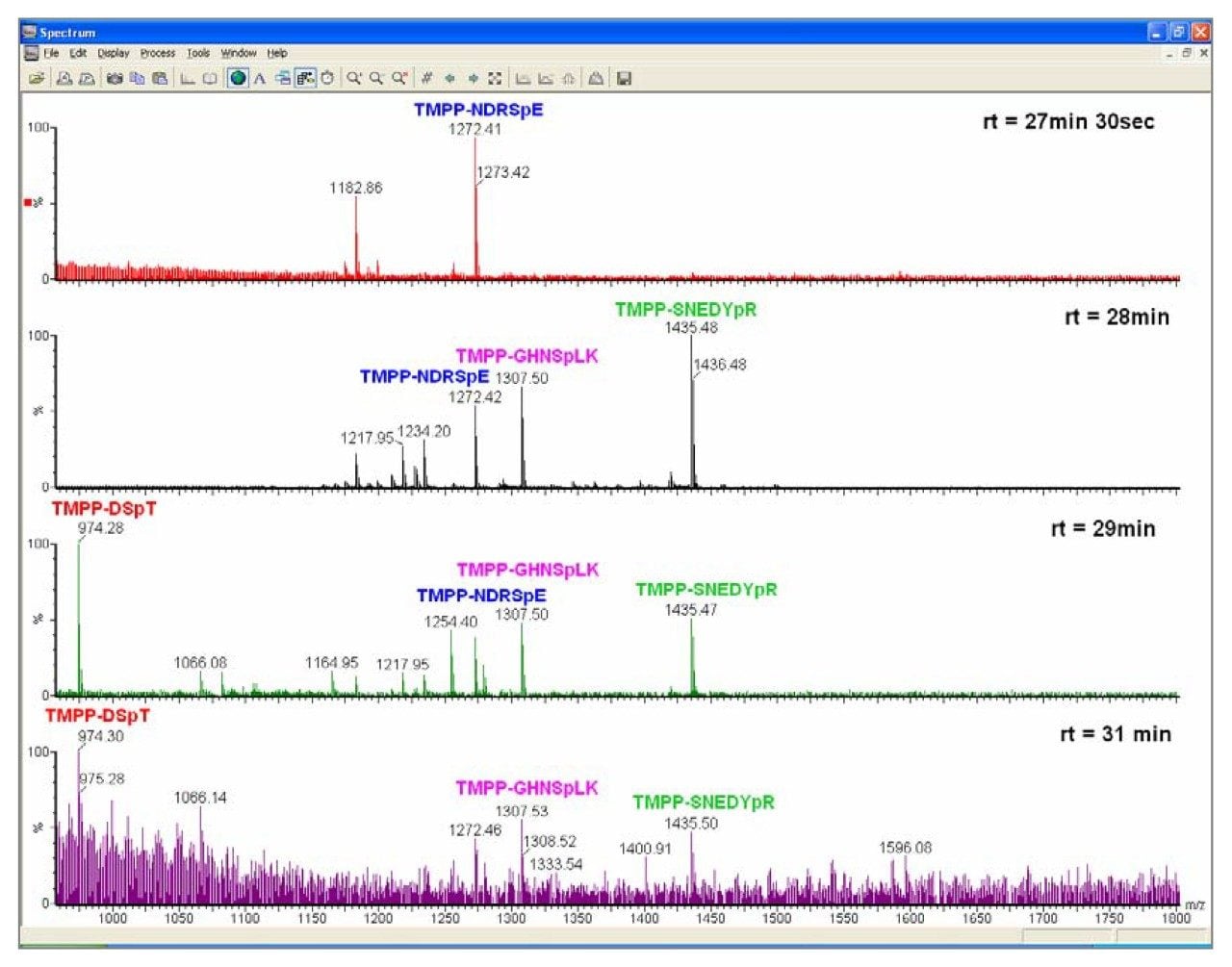
The order of elution is slightly different in the LC/ESI and LC/MALDI experiments. The range of retention times between species was similar in both experiments. Reasons for the difference in chromatography observed are the slightly different chromatographic conditions used. In the LC/ESI experiment the aqueous mobile phase has 1% formic acid, whereas in the LC/MALDI experiment the aqueous mobile phase was 0.1% TFA.
720001308, August 2005