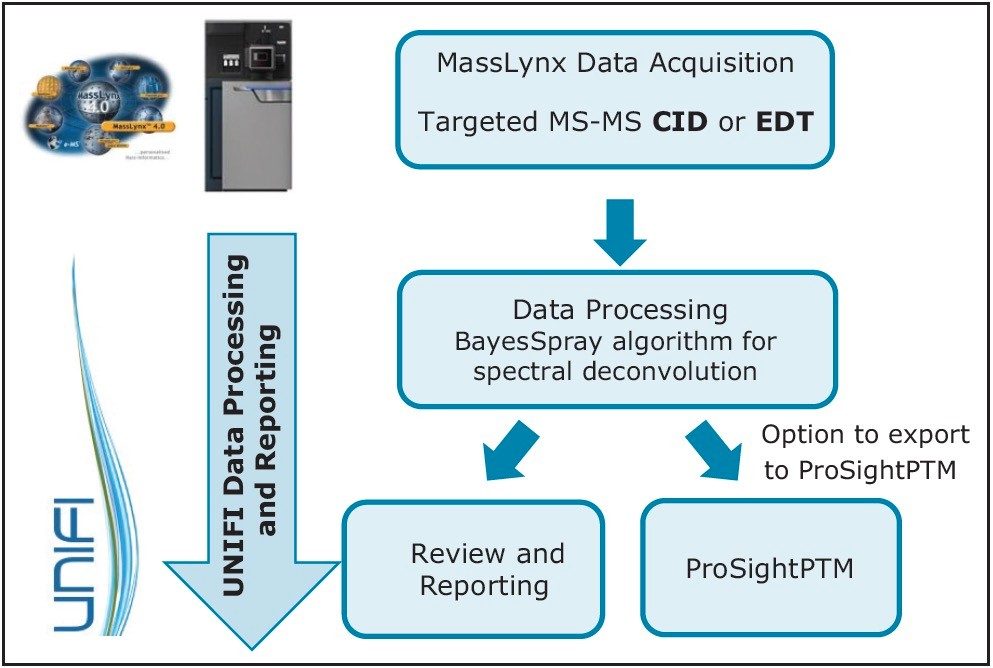
This application note demonstrates the acquisition of protein top-down analysis data by collision induced dissociation (CID) or electron transfer dissociation (ETD) MS-MS fragmentation from the SYNAPT G2-Si HDMS Mass Spectrometer.
The UNIFI Scientific Information System streamlines data processing, reviewing, and reporting for monoclonal antibody fragmentation data, and enables the confirmation of protein sequences and post-translational modifications.
Direct fragmentation of proteins (top-down) provides an orthogonal approach to verify terminal sequence confirmation and localize protein modifications. Protein fragmentation approaches are primarily qualitative and have limited dynamic range to assess variation. Additionally, these approaches have no simple mechanism to determine modification abundance, making them a complementary technology to peptide mapping for mAbs and other biotherapeutics. Top-down protein fragmentation generates diverse fragment ion types and a multitude of charge states with overlapping spectral patterns. This data complexity complicates the process of producing primary structural assignments – usually through manual annotation – leading to the necessary development of software tools that automatically deconvolute raw spectral MS-MS data, annotate fragmentation patterns to protein sequences, and generate reports. These challenges have impeded broader usage of top-down protein analysis for routine biotherapeutic mAb development.
This application note demonstrates the acquisition of protein top-down analysis data by collision induced dissociation (CID) or electron transfer dissociation (ETD) MS-MS fragmentation from the SYNAPT G2-Si HDMS Mass Spectrometer (Figure 1). The data from these analyses was imported into UNIFI for processing, review, and report generation. This automated data processing workflow enabled efficient sequence verification, along with identification of modifications for a mAb and its subunits.
Trastuzumab was treated by IdeS (Genovis) protease and reduced by DTT (SigmaAldrich) to produce light chain (LC), Fd, and Fc/2 fragments. Intact mass and the subsequent top-down analyses were performed on an ACQUITY UPLC H-Class Bio System connected to a SYNAPT G2-Si HDMS Mass Spectrometer. The analyses were processed in UNIFI using the dedicated protein intact mass and top-down workflows. Data acquisition for the CID or ETD MS-MS fragmentation was performed on selected charge states.
|
LC system: |
ACQUITY UPLC H-Class Bio System |
|
Detector: |
ACQUITY UPLC Tunable UV (TUV), 280 nm |
|
Column: |
ACQUITY UPLC Protein BEH C4, 1.7 μm, 2.1 mm x 50 mm (P/N 186004495) |
|
Column temp.: |
80 °C |
|
Sample temp.: |
4 °C |
|
Mobile phase A: |
0.1% formic acid |
|
Mobile phase B: |
0.1% formic acid in acetonitrile |
|
Time(min) |
Flow rate (mL/min) |
%A |
%B |
Curve |
|---|---|---|---|---|
|
Initial |
0.4 |
95 |
5 |
Initial |
|
1.00 |
0.4 |
95 |
5 |
6 |
|
1.10 |
0.1 |
95 |
5 |
6 |
|
2.50 |
0.1 |
77 |
23 |
6 |
|
12.50 |
0.1 |
71 |
29 |
6 |
|
13.00 |
0.1 |
5 |
95 |
6 |
|
13.10 |
0.4 |
5 |
95 |
6 |
|
14.00 |
0.4 |
5 |
95 |
6 |
|
14.50 |
0.4 |
95 |
5 |
6 |
|
17.00 |
0.4 |
95 |
5 |
6 |
|
MS system: |
SYNAPT G2-Si HDMS |
|
Mode: |
ESI+ sensitivity mode |
|
Capillary: |
2 kV |
|
Sample cone voltage: |
40 V |
|
Source temp.: |
120 °C |
|
Desolvation temp.: |
500 °C |
|
Desolvation gas flow: |
800 L/h |
|
Full scan MS: |
Scan rate=0.5 sec Mass range=500–2000 m/z |
Scan rate=1 Hz; mass range=50–2000 m/z
For CID fragmentation, the collision energy level was ramped from 20 to 40 eV
For ETD fragmentation, MS-MS data was acquired over a one second scan (signal accumulation) period with an anion refill time of 100 ms between scans
MassLynx v4.1 for SYNAPT instrument control and data acquisition
UNIFI Scientific Information System v1.8 for data processing, reviewing, and reporting1
ProSight PTM 2.0 as an alternative informatics annotation engine using data exported from UNIFI in the .PUF file format2
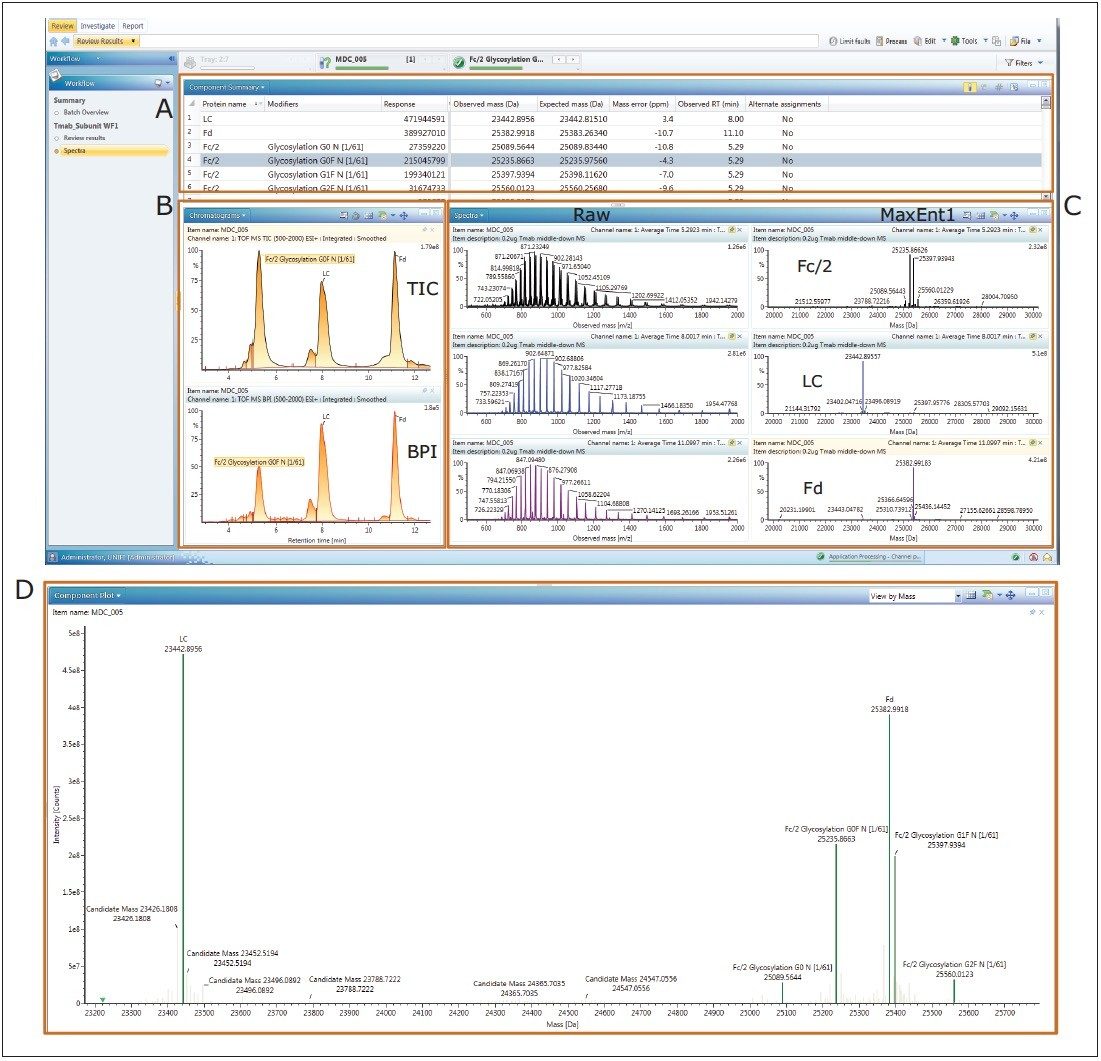
Trastuzumab subunit LC-MS analysis data was acquired on an ACQUITY UPLC H-Class Bio System coupled with a SYNAPT G2-Si HDMS Mass Spectrometer under MassLynx control. The data was then imported into UNIFI for processing, review, and report generation. The UNIFI review panel (Figure 2) includes: (A) A component summary table displaying the trastuzumab subunit and assigned glycoforms; (B) Total ion count (TIC) and base peak ion (BPI) chromatograms; (C) Summed raw MS spectra and MaxEnt1 deconvoluted spectra; (D) Component plot.
The T-mab subunits and their modified forms were identified. The light chain (LC), Fd, and Fc/2 glycoforms (G0, G0F, G1F, and G2F) were displayed with the deconvoluted average mass in the component table (Figure 2A) and the component plot (Figure 2D). The TIC chromatogram exhibited good separation of the subunit peaks with identity annotation (Figure 2B). Easy access to the raw and deconvoluted spectra facilitated data inspection and reviewing (Figure 2C).
Following the subunit analysis, a highly-charged precursor ion was chosen for efficient ETD fragmentation with automated data processing and reporting.
UNIFI provides a new protein top-down analysis workflow (Figure 3) featuring a novel Bayesian inference algorithm – BayesSpray – for deconvolution of both isotope resolved (peptide) and non-resolved (protein) data. It can automate the processing and reporting of collision induced dissociation (CID) as well as electron transfer dissociation (ETD) MS-MS fragmentation, enabling streamlined sequence verification and identification of modification sites.

An example data of infusion top-down analysis with ETD fragmentation on Fc/2 subunit is shown in Figure 4. One precursor ion (765.8 m/z, 33+) corresponding to the Fc/2 G0F glycoform was selected for ETD fragmentation. The MassLynx raw data was then imported into UNIFI for automated processing. The review panel display can be easily customized and configured to interactively investigate the processed data. As an example, an intuitive data review panel is displayed in Figure 4: (A) Component summary table to display the identified trastuzumab subunit; (B) Sequence coverage map; (C) Summed raw, BayesSpray mocked, and deconvoluted CID fragmentation spectra; (D) Annotated deconvoluted spectra and fragmentation ion table. 42 c- and 41 z-ions were assigned, which is roughly 38.1% backbone fragment ion coverage for the Fc/2 subunit.

The UNIFI top-down workflow offers an option to export the processed MS-MS data (BayesSpray deconvoluted fragment ions/intensity list) in a .PUF format file that can be processed further with ProSight. ProSight was designed by the Proteomics Center of Excellence at Northeastern University for protein top-down analysis in order to identify fragments and obtain sequence coverage information.
The example result from ProSight PTM 2.0 assigned the same fragment ions and had the same backbone fragment ion coverage as the UNIFI results (Figure 5). However, the amino acid modifications must be assigned manually to be considered when using ProSight, whereas a UNIFI analysis method can include multiple modifications. In this case, ProSight PTM highlighted N in orange, which was assigned with a custom modification by manually adding the mass of G0F.

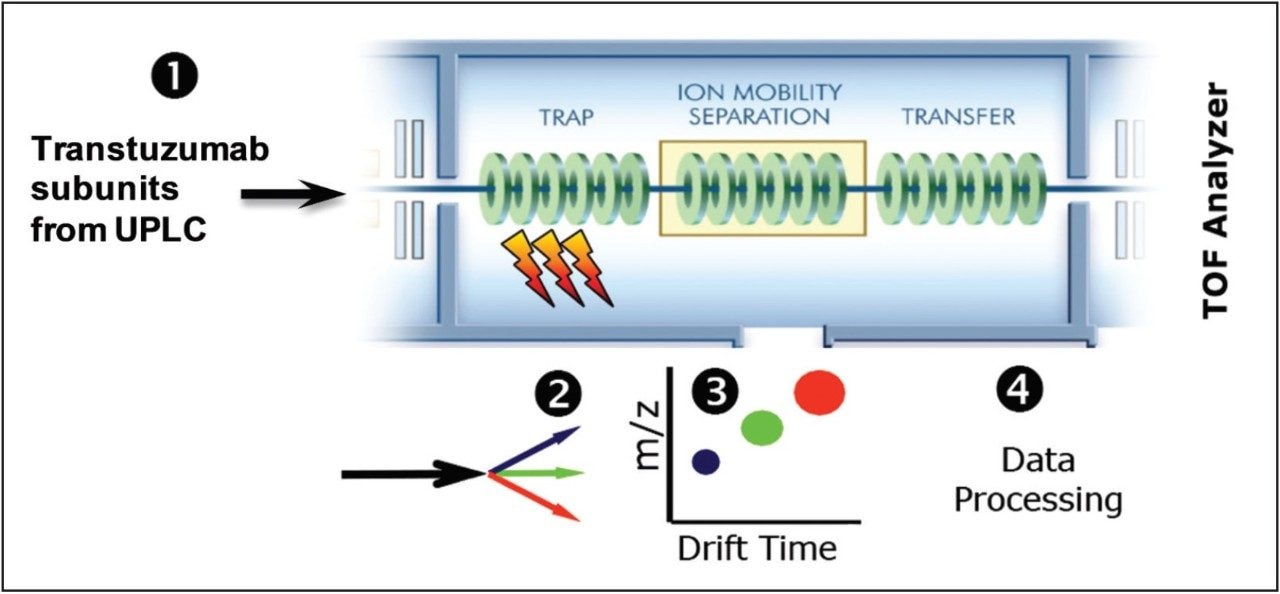
Beyond the conventional top-down methodology, the SYNAPT G2-Si HDMS Mass Spectrometer provides a unique capability for postfragmentation ion mobility separation. This additional gas phase separation of the fragment ions is based on the charge and collision cross sectional differences (Figure 6).
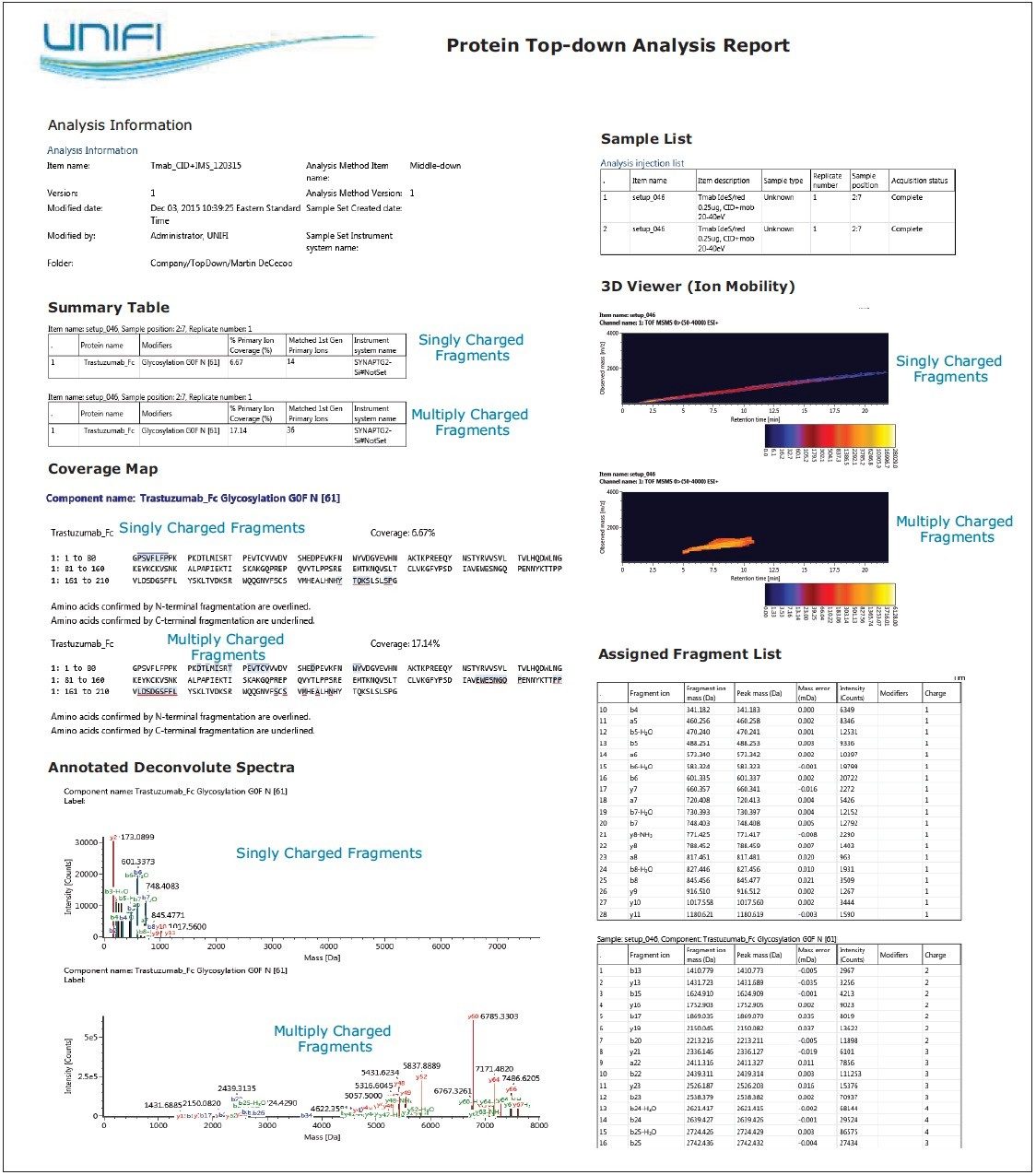
The same sample was separately analyzed by LC-MS/MS (CID-IMS) on SYNAPT G2-Si HDMS. Singly and multiply charged fragment ions were extracted into separate raw files by DriftScope prior to the data being imported into UNIFI and processed. Figure 7 shows the Fc/2 G0F (precursor m/z 902.2, 28+) top-down analysis data with CID fragmentation followed by ion mobility separation. Figure 7B and 7C display the results derived from isolated, singly charged and multiply charged fragments. The raw spectra with only the 1+ charged ions facilitated the manual data interpretation. The fragments were assigned exclusively to the N- and C- terminal sequences. In total, 19 b- and 31 y-ions were assigned, which is roughly 23.81% fragment ion coverage for the Fc/2 subunit. This additional separation produced more intuitive spectra, and simplified data review and terminal sequence confirmation.

Report templates can be created and customized to meet specific requirements, and then saved to be applied to future analyses. Figure 8 provides a snapshot of the trastuzumab Fc/2 CID-IMS top-down analysis report of extracted singly and multiply charged fragment ions. This report includes acquisition and sample information, sequence coverage map, fragmentation viewer/table, and 3D illustration of IMS data – all of which can be organized within a single report format. The object properties and report templates are user-configurable. Multiple reports can be executed for one analysis to answer all scientific questions in an efficient format.
The described automated workflows for intact mass and top-down protein analysis facilitated the confirmation of protein terminal sequences and localization of modifications suggested by the intact mass results. This data processing workflow supported MS-MS data acquired with either CID or ETD fragmentation, overcoming the challenges of manually interpreting and reporting complex protein top-down results. The SYNAPT G2-Si HDMS System provided the unique capability toseparate fragment ions by gas-phase ion mobility, generating simplified spectra for inspection and reviewing. Extracting and analyzing singly charged fragment ions from the top-down experiment enabled the terminal sequence to be readily confirmed, while multiply charged fragments provided data of the internal regions of the sequences within the mAb subunit. This ability to automate processing, fragment assignment, and organization of results in a templated UNIFI report facilitates the efficient communication of top-down data within and across biopharmaceutical organizations.
720005689, May 2016