This is an Application Brief and does not contain a detailed Experimental section.
This application brief establishes that the BioAccord System exhibits sufficient reproducibility for peptide mapping in regulated workflows subject to traditional validation requirements.
Inter- and intra-system comparison of the peptide mapping workflow using the BioAccord System.
In 2017, the FDA published a paper titled, “A Retrospective Evaluation of the Use of Mass Spectrometry in FDA Biologics License Applications”,¹ which highlights the increased use of mass spectrometry for characterization of protein-based biotherapeutics. Most recently, there is a concerted effort from the pharmaceutical industry to extend the use of liquid chromatography and high-resolution mass spectrometry (LC-MS) beyond characterization, particularly for attribute monitoring analysis within late stage development and QC environments. Concerns exist regarding the need for highly skilled scientists to generate and collect this data, and for robustness and variability of LC-MS platforms for developing MS-based assays in process development and QC laboratories. The launch of the Waters BioAccord System represents the first built-for-purpose commercial platform to address the industry’s needs for a robust, compliance-ready, and easily deployable high-performance LC-MS system for biopharmaceutical analysis. As illustrated in a myriad of application and technical notes,1-9 the BioAccord System meets and exceeds operational requirements in these areas. In this latest study, we focused on the reproducibility of the BioAccord System for the peptide mapping workflow, complementing earlier work on intact mass analysis (Waters Application Note, 720006722).
The BioAccord System is comprised of an ACQUITY UPLC I-Class PLUS System configured with optical detection (TUV or FLR) and coupled in-line with the ACQUITY RDa Mass Detector (a compact high-performance oa-TOF Mass Detector). The system is operated under the waters_connect platform, with the UNIFI Scientific Information System providing streamlined workflows for automated data acquisition, processing, and reporting of peptide map, intact mass, released glycan, and oligonucleotide analysis. This system is purposely designed for routine biopharmaceutical analysis, deployable in both regulated and non-regulated laboratories.
Peptide mapping is used routinely to confirmtherapeutic protein primary sequences and establish patterns, as well as levels of product modification variants. Reliable analysis of lower abundance variants is very sensitive to both sample handling and LC-MS instrument performance. Therefore, robust, easy-to-use, and reproducible LC-MS systems and methods are critical for developing peptide mapping assays that can be validated within regulated environments.
In this study, the Waters mAb Tryptic Digest Standard (p/n: 186009126) was used for all peptide mapping experiments and 200 μL of water were added to the sample vial (which contains 40 μg of tryptic digested NISTmAb) for a final concentration of 0.2 μg/μL. Five microliters (5 μL) of the solution were injected into the BioAccord System for analysis (1 μg on column injection). A Waters ACQUITY UPLC BEH C18 130 Å, 1.7 μm, 2.1 mm X 100 mm Column (p/n: 186002352) was heated to 65 °C for the peptide separation. Mobile phases A and B were 0.1% formic acid in milli-Q water and 0.1% formic acid in acetonitrile respectively. A linear gradient of 99% A to 60% A over 60 min was established at 0.2 mL/min flow rate. The total run time was 75 min for the analysis, including column equilibration. The RDa Detector was operated in the ESI positive full scan with fragmentation mode (at 2 Hz) with data acquired over the mass range of 50–2000 m/z. The cone voltage was 30 V (with fragmentation cone voltage ramped from 60 V to 130 V), capillary voltage was set to 1.5 V and the desolvation temperature was kept at 500 ºC. Intelligent data capture was enabled, and the mass tolerance for peptide assignment was set at 10 ppm with a minimum number of confirmatory fragment ions >3 as an additional filter to minimize false assignments. The waters_connect “quantify assay Tof 2D chromatographic” analysis type within the UNIFI app was used to quantify the relative abundance of modified peptides.
Figure 1 shows the overlay of the total ion chromatograms (TICs) from 18 peptide injections on a BioAccord System, over about 22 hours run time. The overlay of the 18 peptide map chromatograms shows retention time reproducibility with 0.09 minutes or less standard deviation for all monitored peptides, as also shown in Table 1.
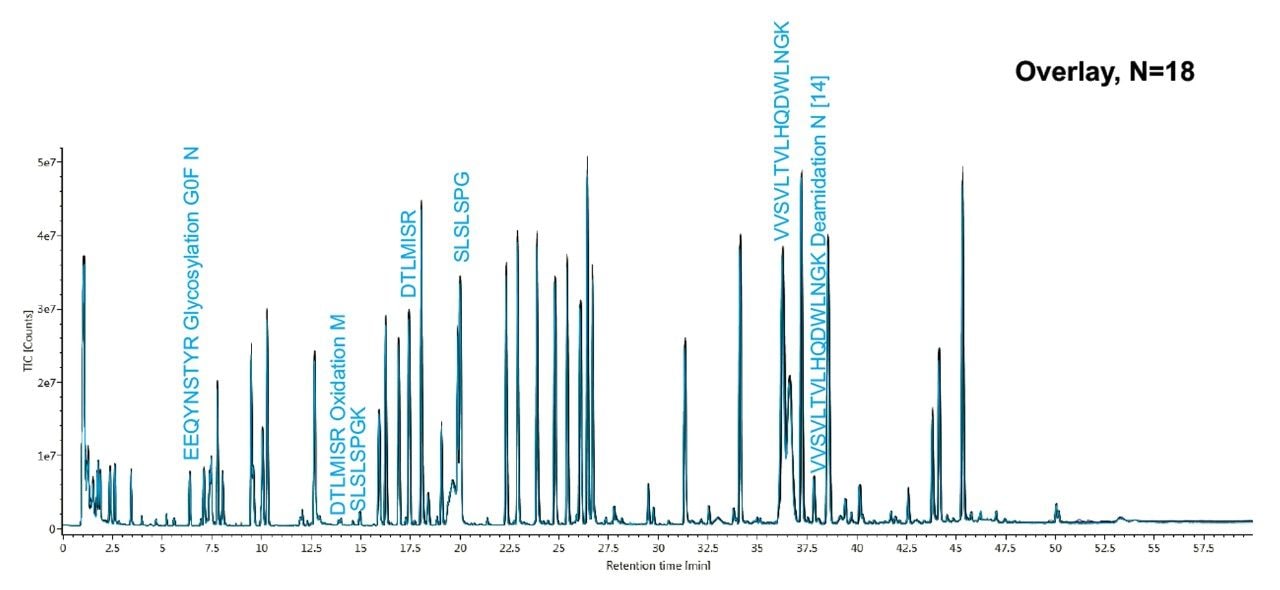

To demonstrate the BioAccord System performance for reliable detection of peptide modification levels, a set of native peptides and their modified counterparts were targeted for relative quantification within the analysis.
These peptides include glycopeptide, EEQYNSTYR with G0F, G1F and G2F, DTLMISR (Oxidation M), and VVSVLTVLHQDWLNGK (Deamidation N), as labeled in Figure 1.
The same LC-MS analysis was conducted with two additional BioAccord systems (18 injections), with comparable peptide retention times across the three systems. It was found that the retention time standard deviation for all monitored peptides was 0.09 minutes (5.4 seconds) or less between the three systems.
Plotting the relative abundances of the same set of peptides using the data sets generated across the three systems (Figure 2) indicates some small inter-system fluctuation of the relative abundance for some of the glycopeptides. However calculated %RSD for all the peptides monitored is within 8.12% across peptides of significantly different MS response.
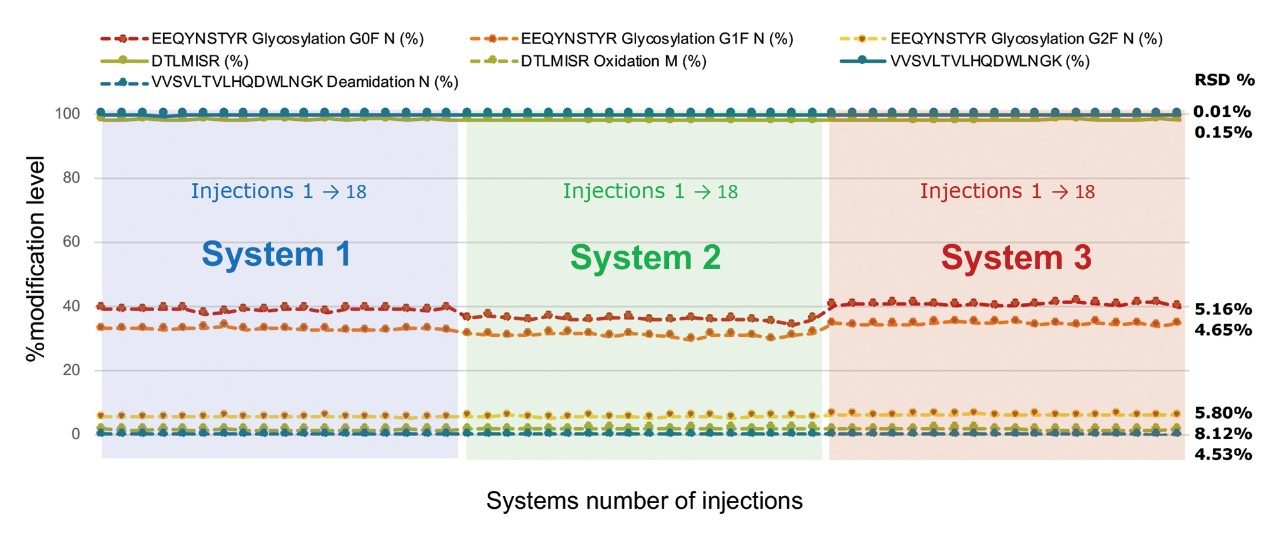
The inter- and intra-system studies show that the BioAccord systems are fit-for-purpose for peptide mapping analysis within regulated environments based on data generated from 18 injections each on three systems, The retention time standard deviation of all monitored peptides was less than 0.09 minutes, and the relative abundance of all monitored peptide modifications exhibited an average %RSD within 8.12% across varying levels of peptide intensity.
Consistent with our previous study of inter- and intra-system intact mass analysis performance,⁹ peptide mapping data generated from BioAccord systems supports the confident deployment of MS-based assays in late stage development and QC organizations.
720006831, April 2020