This application note demonstrates how BioAccord LC-MS System can be used for peptide mass confirmation and impurity profiling within research, development, and more highly regulated laboratories in manufacturing and QC. The integrated workflows within UNIFI allow accurate mass-based identification, sequence conformation, and relative quantification of biotherapeutic peptides and their impurities on a single platform, with SmartMS functionality making these capabilities accessible to a broad segment of scientists and technicians within the organization.
Today, a wide variety of peptide biotherapeutics are progressing through the development pipeline due to their numerous therapeutic benefits and significant advancements in technologies for synthetic and recombinant production, downstream processing, and bio delivery. While varied in physical properties, most of these molecules have molecular mass below 5000 Da. Ultimately, peptide purity depends upon optimization of various parameters during the production and processing steps, which requires analytical tools capable of assessing these molecules with rapid and reliable results for confirming the main product, any characteristic impurities, and to detect any unexpected product modifications. Generally, conventional liquid chromatography assays with optical detection are utilized to address the impurity profiling, but these assays have certain limitations for unresolved impurities and addressing the cause of newly discovered peaks, which makes the use of more advanced analytical instruments capable of delivering more structural information on these profiles a path for addressing these challenges in quick time.
Mass spectrometry is well suited for establishing the identity and purity of biotherapeutic peptides1. The incorporation of high-resolution mass spectrometry into the analytical workflow can provide accurate mass-based confirmation of peptide API, known impurities, along with providing verification of peptide sequences via their fragment ions2. The BioAccord LC-MS System is a high performance analytical platform that was designed and developed to be efficiently and easily deployed and operated even by laboratories lacking previous experience with LC-MS technologies. The SmartMS capabilities of the BioAccord are manifested with a simplified user interface, automated startup, and advanced self-diagnostics capabilities. This application note demonstrates how the BioAccord System supports an integrated workflow for biotherapeutic peptide analysis and impurity profiling.
Liraglutide, a glucagon-like peptide -1 receptor agonist, is a therapeutic peptide of 31 amino acids (HAEGTFTSDV SSYLEGQAAK EFIAWLVRGR G) with attachment of palmitic acid chain to a lysine side chain via a glutamic acid linker and has a monoisotopic mass of 3748.9465 Da. It can be produced by both rDNA technology and chemical synthesis. Liraglutide of rDNA origin is used in this study to demonstrate integrated workflow on the BioAccord System. The compliance-ready UNIFI application on the waters_connect Informatics platform enabled a streamlined workflow, combining automated data acquisition, processing, and reporting.

A solution of liraglutide (rDNA origin) at 6 mg/mL is used.
|
LC-MS system: |
BioAccord incorporating the ACQUITY RDa Mass Detector ACQUITY UPLC I-Class PLUS and ACQUITY UPLC TUV Detector |
|
Column: |
ACQUITY UPLC Peptide CSH C18, 130 Å, 1.7 µm, 2.1 x 150 mm (p/n: 186006938) |
|
Column temp.: |
45 °C |
|
TUV wave length: |
215 nm |
|
Flow rate: |
0.12 mL/min |
|
Mobile phase A: |
0.1% Formic acid in water |
|
Mobile phase B: |
0.1% Formic acid in acetonitrile |
|
Injection volume: |
1 µL |
|
Steps |
Time (min) |
Solvent A Composition (%) |
Solvent B Composition (%) |
Curve Profile |
|---|---|---|---|---|
|
1 |
0.00 |
95 |
5 |
Initial |
|
2 |
2.00 |
70 |
30 |
6 |
|
3 |
80.00 |
45 |
55 |
6 |
|
4 |
100.00 |
5 |
95 |
6 |
|
5 |
101.00 |
95 |
5 |
6 |
|
6 |
105.00 |
95 |
5 |
6 |
|
Mode: |
Full scan with fragmentation |
|
Mass range: |
50–2000 m/z |
|
Polarity: |
Positive |
|
Capillary voltage : |
1.50 kV |
|
Desolation temp.: |
550 °C |
|
Cone voltage: |
50 V |
|
Fragmentation cone voltage: |
95–100 V |
|
Lockmass: |
Waters_connect Lockmass Solution (p/n: 186009298) |
|
Informatics: |
waters_connect platform with UNIFI Application |
The LC-HRMS strategy for biotherapeutic peptide mass confirmation and impurity profiling utilizes the UNIFI informatics peptide mapping workflow for characterization of the peptide and impurity peaks, and accurate mass screening workflow for targeted impurity profiling and automated relative % measurements. Data from the characterization stage is used to generate a targeted list of peptide species for accurate mass monitoring and quantification.
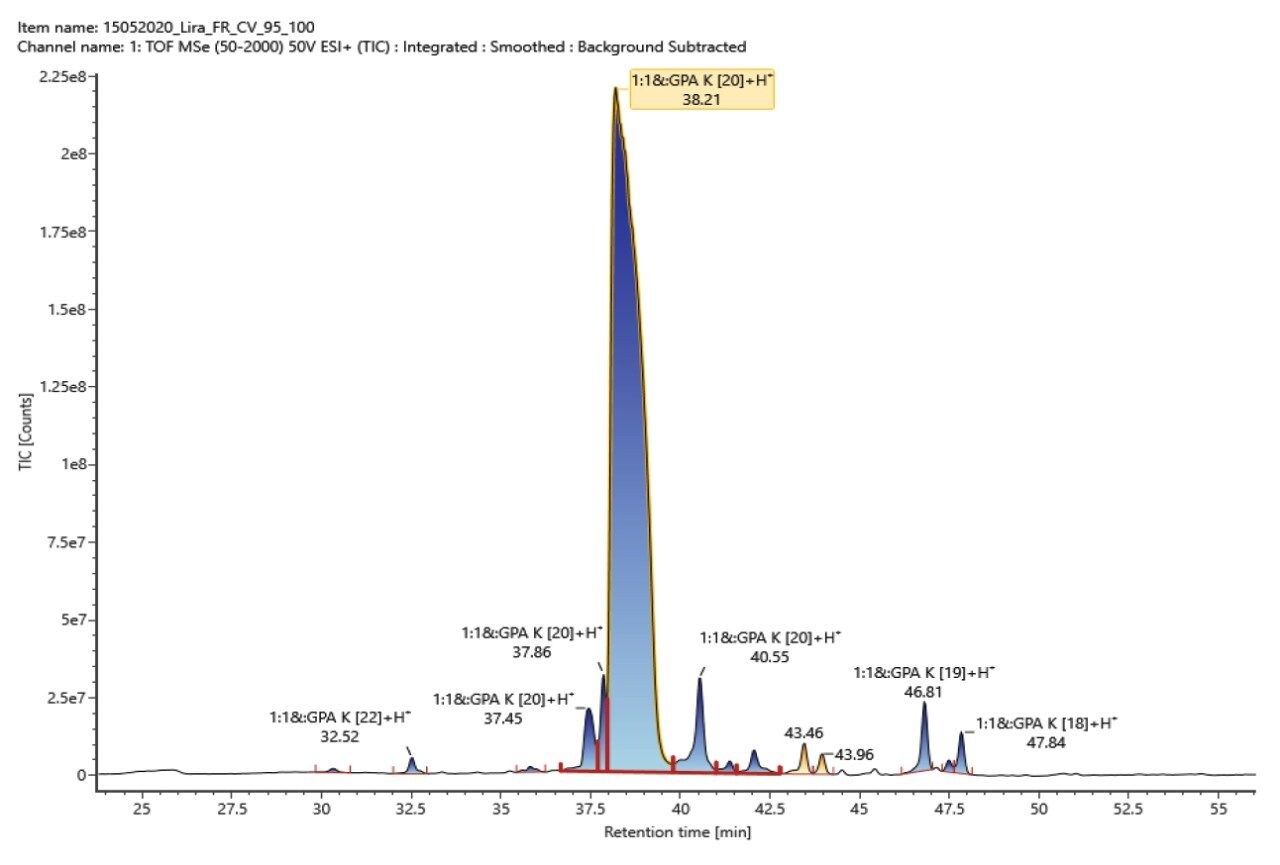
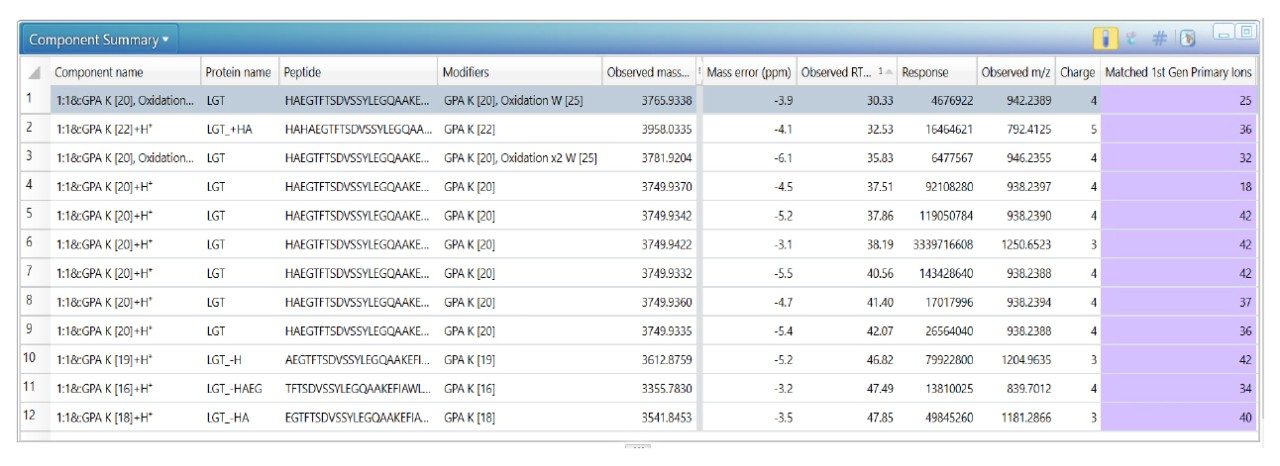
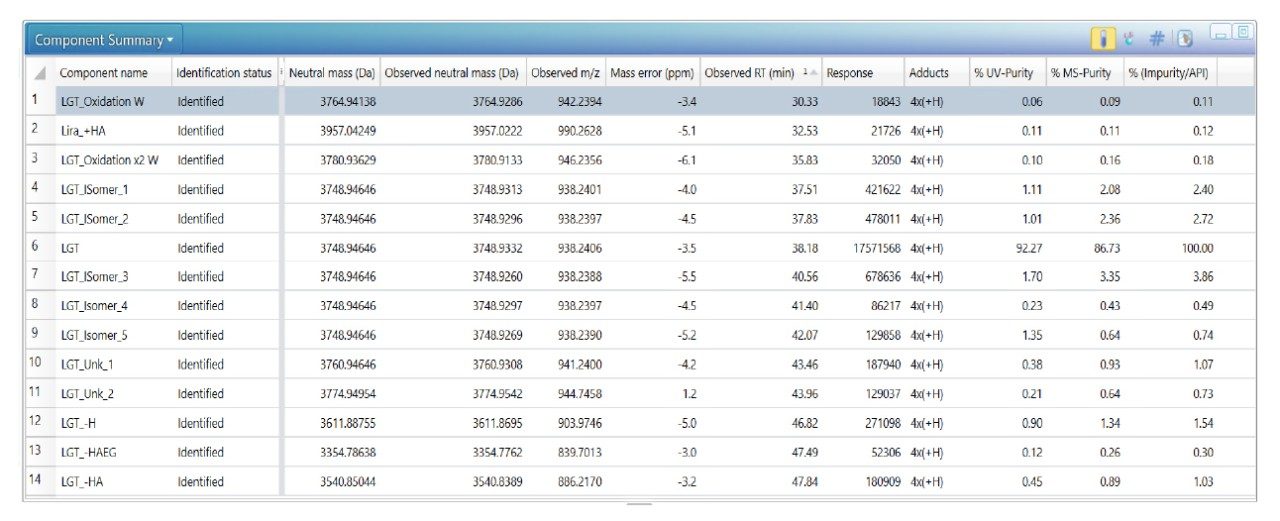
The impurities in liraglutide were first chromatographically separated on an ACQUITY UPLC Peptide CSH C18 130 Å Column using a shallow acetonitrile gradient in 0.1% formic acid. The acquired LC-MS data was then processed by the peptide mapping workflow method to identify the impurities and confirm the peptide sequence of impurities and liraglutide. Figure 2 shows the zoomed total ion chromatogram of the liraglutide sample. Peaks are labeled with respective identifications, and Figure 3 shows the component summary providing details of these identifications. The impurities observed were isomers, N-terminal truncations, additions, and oxidized forms of liraglutide.
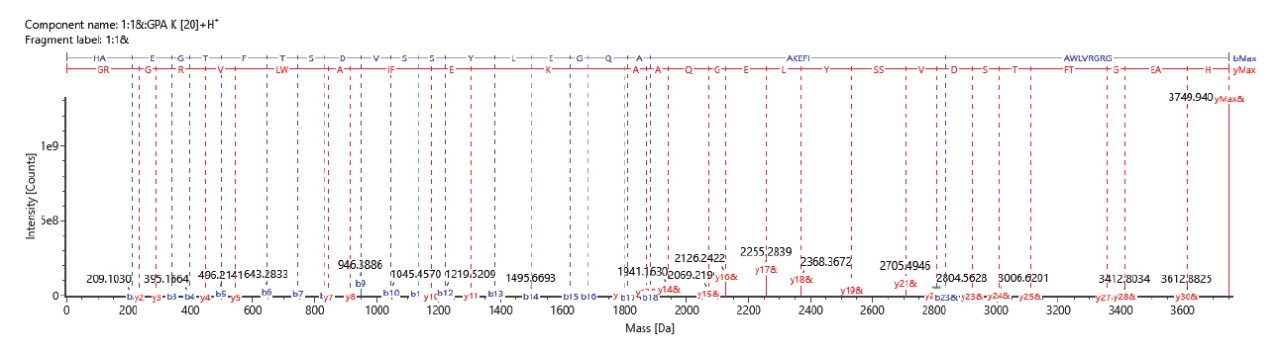
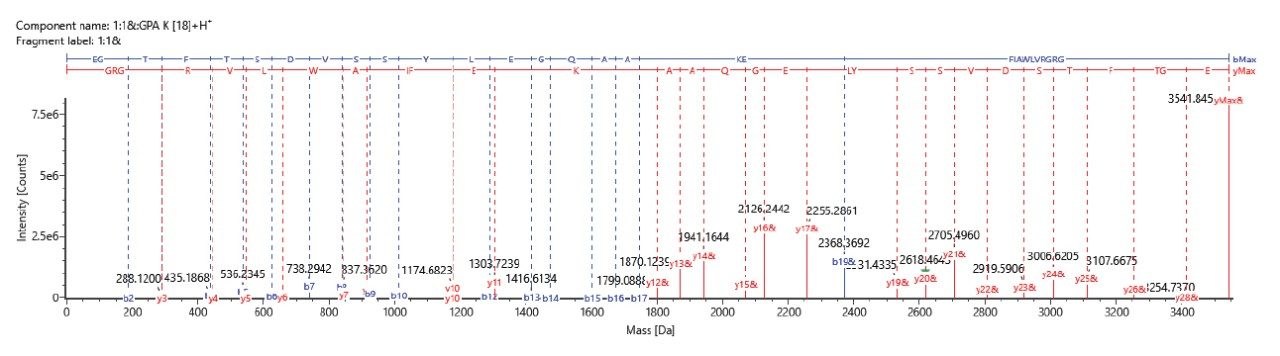
In one of its modes of operation, the ACQUITY RDa is capable of generating structurally informative fragment ions, adding more confidence to accurate mass-based peptide assignments. This unique feature is achieved by altering MS scans (one with lower collisional energy, and one with higher collisional energy ramping) during data acquisition. As a result, the instrument can be operated in MS only (MS 1) or Full scan with fragmentation (data independent acquisition) modes (MS 2) with collision induced fragmentation. Figure 4 and 5 shows the fragmentation spectra for liraglutide containing palmitic acid chain attached to Lys20 sidechain via a glutamic acid linker, and liraglutide truncated impurity (observed at relative level of 0.45% in UV detection) confirming the loss of the N-terminal two amino acids ‘HA’ respectively. The automated data processing generates annotated fragment ions spectra for all detected components.
Existing amino acid modifications can be selected during UNIFI processing to assign peptide impurities. Novel amino acid modifications can be created to supplement the default set of modifications in the software. Also, unnatural amino acids can be created and saved in UNIFI scientific library. The scientific library application has a simple interface that enables the user to generate these new modifications (Figure 6).
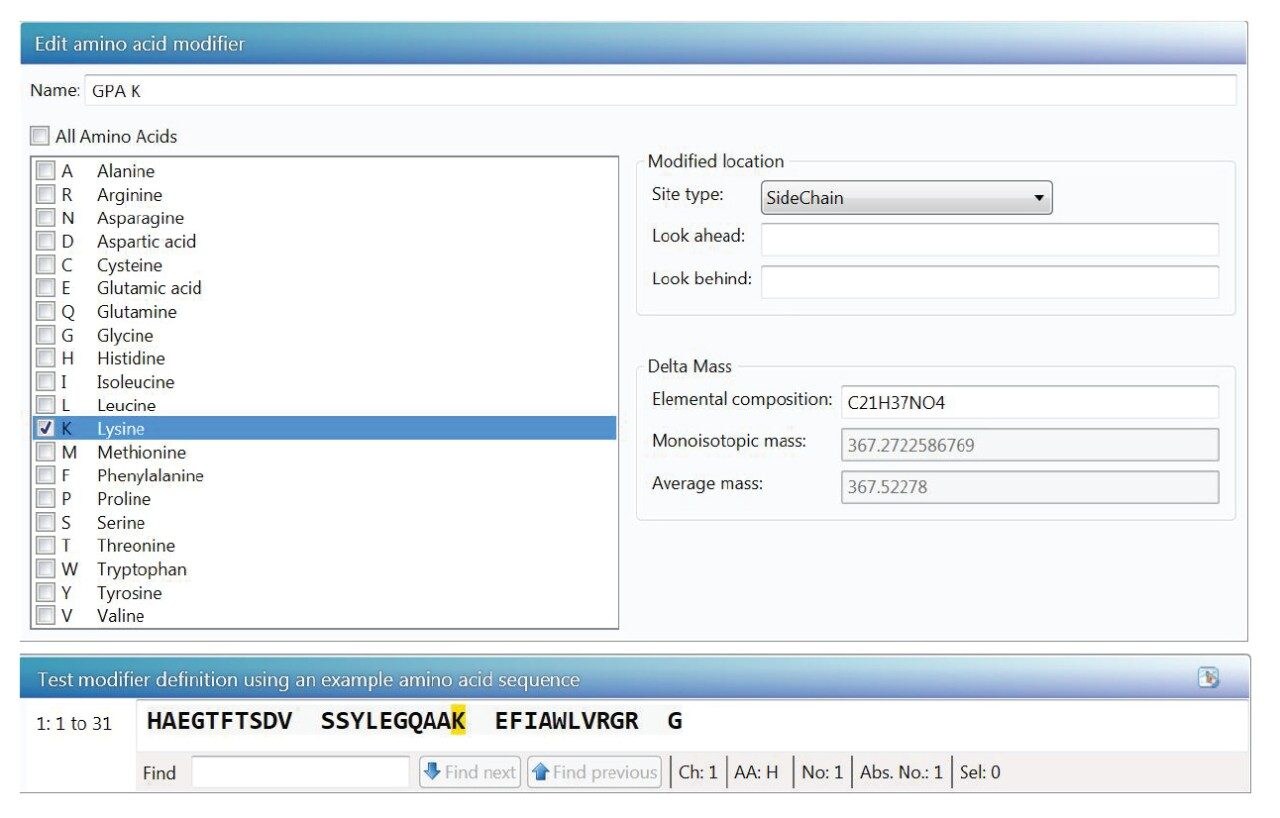
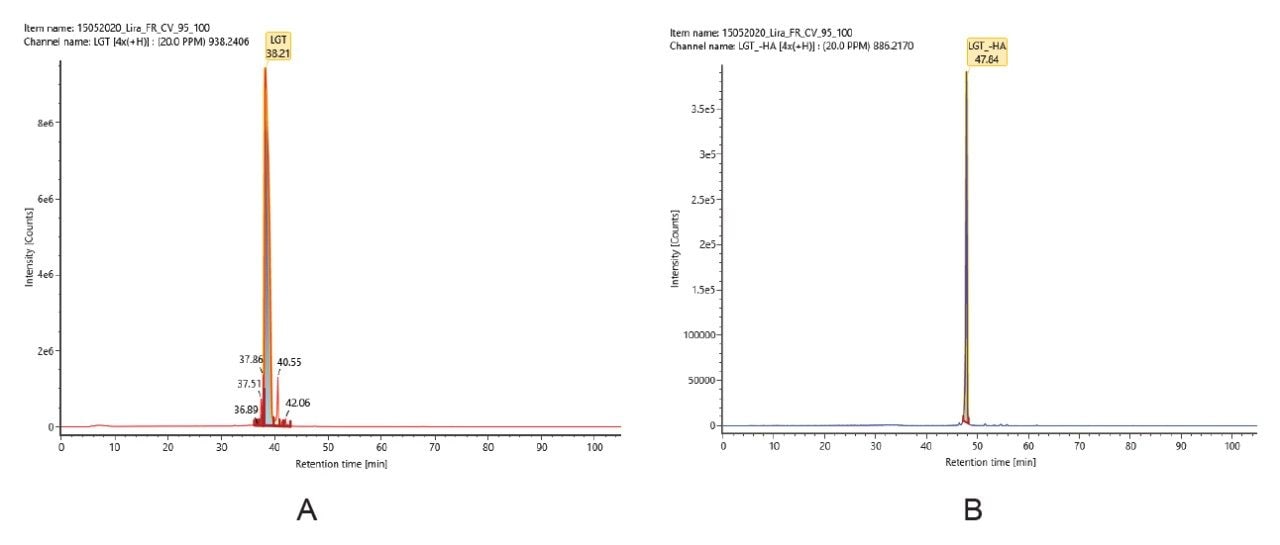
Impurities identified in characterization stage by the peptide mapping workflow can be added to a scientific library file and imported to the accurate mass screening workflow for targeted impurity profiling. Additional library entries can also be created based on prior knowledge. The accurate mass screening workflow uses XIC of each component targeted based on mass and retention time information on this component target list. The accurate mass screening workflow is used to automate the determination of % purity of peptides based on UV and MS response and %relative abundance to main peak MS response. Figure 7 shows the accurate mass screening workflow’s component summary displaying the purity level of liraglutide and impurity components that are monitored using both MS and UV detection. Figure 8 shows the extracted ion chromatogram (XIC) of liraglutide(A) and truncated liraglutide impurity ‘LGT-HA’ (B).
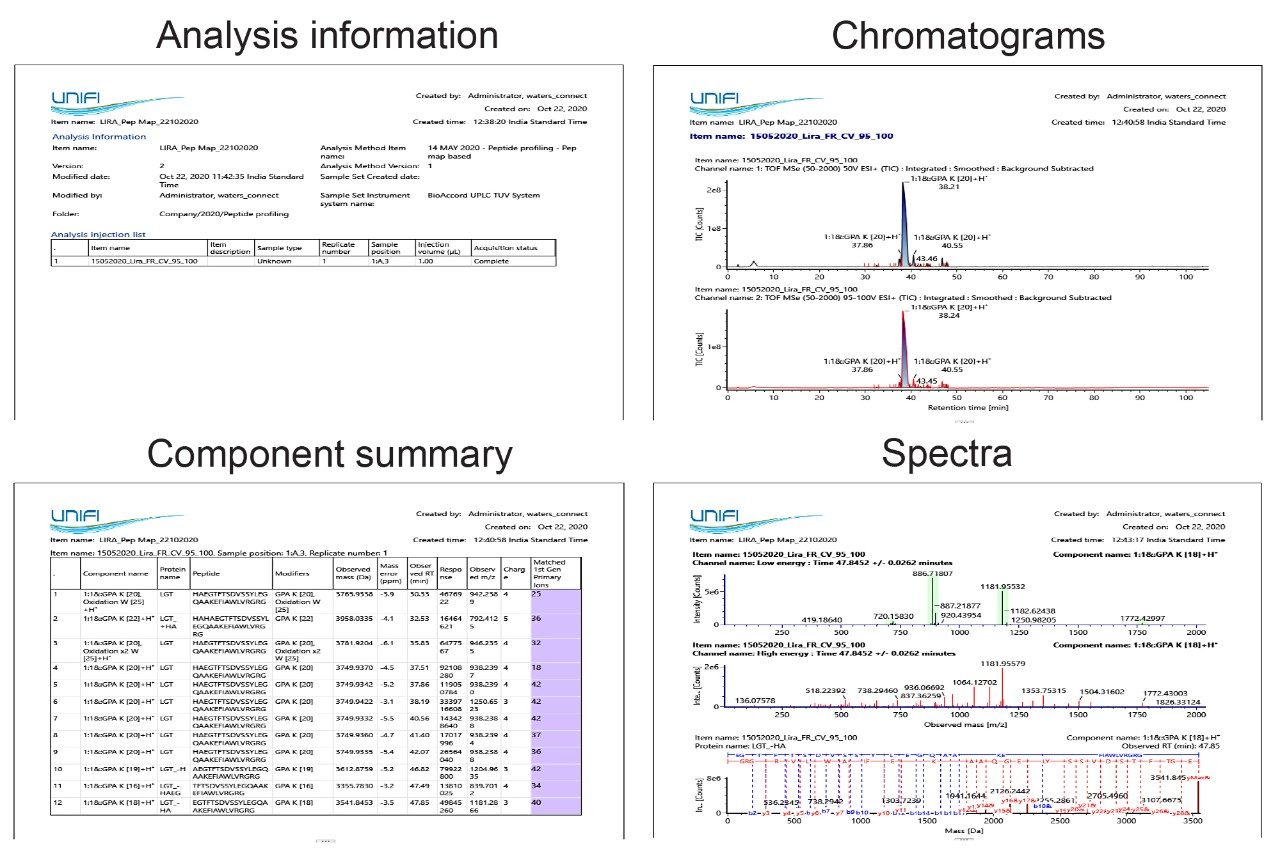
A report or multiple reports can be automatically generated post-acquisition using a pre-defined (default or user-defined) template(s) to summarize the final results of this impurity monitoring study (Figure 9). The software has provisions for electronic signatures on these reports, if necessitated by regulatory or organizational requirements.
Using the peptide mapping and accurate mass screening/monitoring workflows described here with liraglutide, scientists can utilize the BioAccord LC-MS System to perform both mass confirmation and impurity profiling of peptide biotherapeutics in an efficient integrated approach. The significant structural information generated by the addition of the accurate mass data will facilitate more rapid method development, method transfer, and investigations of product deviations, enabling better and faster decision making within peptide biotherapeutic organizations.
This application note demonstrates how BioAccord LC-MS System can be used for peptide mass confirmation and impurity profiling within research, development, and more highly regulated laboratories in manufacturing and QC. The integrated workflows within UNIFI allow accurate mass-based identification, sequence conformation, and relative quantification of biotherapeutic peptides and their impurities on a single platform, with SmartMS functionality making these capabilities accessible to a broad segment of scientists and technicians within the organization.
720007093, December 2020